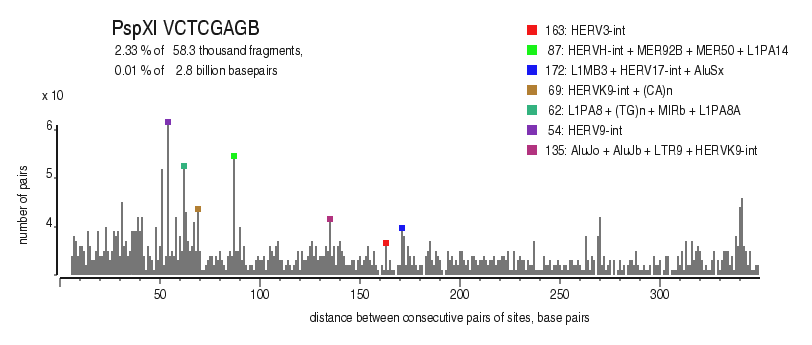
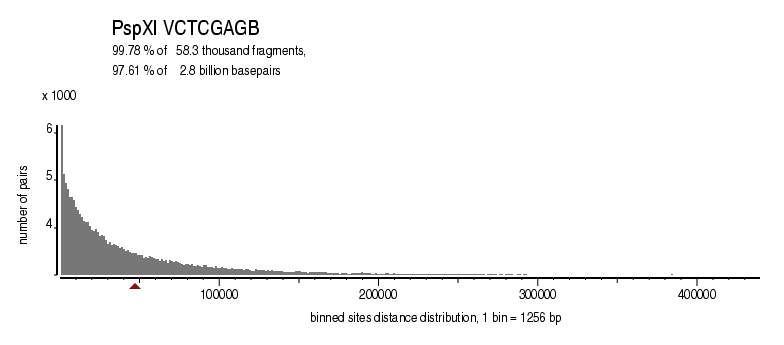
| Enzyme: | PspXI | Longest uncut segments |
| Specificity: | VCTCGAGB | Repeats in uncut segments |
| Number of sites: | 58338 | Genes in uncut segments |
| Mean distance between sites: | 49047 base pairs | |
| Standard deviation: | 62813 base pairs | |
| Site density | 20.4 per megabase | Help |
| Distribution of closely spaced sites |
|---|
 |
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 957095 | chr8 | NT_008183.19 | 35312516-36269611 | 54.81 % in | 1522 repeats | 0.09 % in | 1 genes |
| 2 | 897627 | chr18 | NT_010966.14 | 20912329-21809956 | 45.95 % in | 1464 repeats | 14.06 % in | 1 genes |
| 3 | 890742 | chr2 | NT_005403.17 | 36621487-37512229 | 60.51 % in | 1450 repeats | 11.19 % in | 7 genes |
| 4 | 779609 | chr2 | NT_022184.15 | 56211552-56991161 | 51.72 % in | 1303 repeats | 46.21 % in | 3 genes |
| 5 | 716601 | chr4 | NT_016354.19 | 36740957-37457558 | 45.60 % in | 998 repeats | 0.05 % in | 1 genes |
| 6 | 710297 | chr14 | NT_026437.12 | 63121654-63831951 | 50.76 % in | 1209 repeats | 0.62 % in | 3 genes |
| 7 | 682799 | chr15 | NT_037852.6 | 1230879-1913678 | 16.33 % in | 431 repeats | 2.95 % in | 4 genes |
| 8 | 667271 | chrX | NT_011681.16 | 134453-801724 | 69.23 % in | 1153 repeats | 1.48 % in | 3 genes |
| 9 | 665026 | chr1 | NT_032977.9 | 76332154-76997180 | 58.73 % in | 1002 repeats | 0.00 % in | 0 genes |
| 10 | 663076 | chr2 | NT_022135.16 | 16461328-17124404 | 60.65 % in | 1074 repeats | 0.00 % in | 0 genes |
| 11 | 661768 | chr3 | NT_005612.16 | 1472389-2134157 | 53.71 % in | 1039 repeats | 0.00 % in | 0 genes |
| 12 | 660812 | chr11 | NT_167190.1 | 445881-1106693 | 52.39 % in | 947 repeats | 0.00 % in | 0 genes |
| 13 | 640206 | chr10 | NT_008705.16 | 20385943-21026149 | 50.49 % in | 992 repeats | 0.00 % in | 0 genes |
| 14 | 635342 | chr1 | NT_032977.9 | 42585971-43221313 | 50.22 % in | 984 repeats | 0.00 % in | 0 genes |
| 15 | 633274 | chr4 | NT_016354.19 | 5038406-5671680 | 47.75 % in | 923 repeats | 0.00 % in | 0 genes |
| 16 | 631560 | chr3 | NT_022517.18 | 34775220-35406780 | 53.71 % in | 1046 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 957095 | chr8 | NT_008183.19 | 35312516-36269611 | 1522 | 332 | AT_rich (177) | L2a (45) | (TA)n (43) | |
| 2 | 897627 | chr18 | NT_010966.14 | 20912329-21809956 | 1464 | 318 | AT_rich (110) | MIR (62) | MIRb (61) | |
| 3 | 890742 | chr2 | NT_005403.17 | 36621487-37512229 | 1450 | 346 | AT_rich (146) | L2a (38) | (TA)n (29) | |
| 4 | 779609 | chr2 | NT_022184.15 | 56211552-56991161 | 1303 | 298 | AT_rich (157) | MIR (45) | L2a (43) | |
| 5 | 716601 | chr4 | NT_016354.19 | 36740957-37457558 | 998 | 239 | AT_rich (127) | L2a (49) | MIRb (40) | |
| 6 | 710297 | chr14 | NT_026437.12 | 63121654-63831951 | 1209 | 312 | AT_rich (72) | MIR (59) | MIRb (53) | |
| 7 | 682799 | chr15 | NT_037852.6 | 1230879-1913678 | 431 | 139 | AluSx (25) | AluJb (16) | MIRb (14) | |
| 8 | 667271 | chrX | NT_011681.16 | 134453-801724 | 1153 | 301 | AT_rich (47) | L2a (44) | MIRb (38) | |
| 9 | 665026 | chr1 | NT_032977.9 | 76332154-76997180 | 1002 | 258 | AT_rich (112) | L2a (38) | (TA)n (30) | |
| 10 | 663076 | chr2 | NT_022135.16 | 16461328-17124404 | 1074 | 281 | AT_rich (57) | MIRb (50) | MIR (34) | |
| 11 | 661768 | chr3 | NT_005612.16 | 1472389-2134157 | 1039 | 279 | AT_rich (155) | AluSx (36) | AluY (29) | |
| 12 | 660812 | chr11 | NT_167190.1 | 445881-1106693 | 947 | 241 | AT_rich (102) | MIR (34) | (TA)n (26) | |
| 13 | 640206 | chr10 | NT_008705.16 | 20385943-21026149 | 992 | 245 | AT_rich (63) | AluSx (48) | L2a (37) | |
| 14 | 635342 | chr1 | NT_032977.9 | 42585971-43221313 | 984 | 235 | AT_rich (112) | MIRb (44) | L2a (42) | |
| 15 | 633274 | chr4 | NT_016354.19 | 5038406-5671680 | 923 | 254 | AT_rich (88) | MIRb (43) | MIR (42) | |
| 16 | 631560 | chr3 | NT_022517.18 | 34775220-35406780 | 1046 | 251 | AT_rich (78) | MIRb (48) | MIR (37) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 957095 | chr8 | NT_008183.19 | 35312516-36269611 | LOC100420845 | |
| 2 | 897627 | chr18 | NT_010966.14 | 20912329-21809956 | PIK3C3 | phosphatidylinositol_3-kinase_catalytic_subunit_type_3 |
| 3 | 890742 | chr2 | NT_005403.17 | 36621487-37512229 | ELF2P4 | |
| FSIP2 | fibrous_sheath-interacting_protein_2 | |||||
| LOC100420895 | ||||||
| RPL21P32 | ||||||
| LOC100420976 | ||||||
| RPL23AP35 | ||||||
| LOC100131051 | ||||||
| 4 | 779609 | chr2 | NT_022184.15 | 56211552-56991161 | LRRTM4 | leucine-rich_repeat_transmembrane_neuronal_protein_4_isoform_b |
| TRNAP25P | ||||||
| RPL38P2 | ||||||
| 5 | 716601 | chr4 | NT_016354.19 | 36740957-37457558 | RPL36AP23 | |
| 6 | 710297 | chr14 | NT_026437.12 | 63121654-63831951 | EEF1A1P2 | |
| RPL9P6 | ||||||
| EIF3LP1 | ||||||
| 7 | 682799 | chr15 | NT_037852.6 | 1230879-1913678 | LOC100287945 | |
| LOC283804 | ||||||
| LOC727914 | ||||||
| LOC100418897 | ||||||
| 8 | 667271 | chrX | NT_011681.16 | 134453-801724 | LOC100129229 | |
| CYCSP44 | ||||||
| SPANXN1 | sperm_protein_associated_with_the_nucleus_on_the_X_chromosome_N1 | |||||