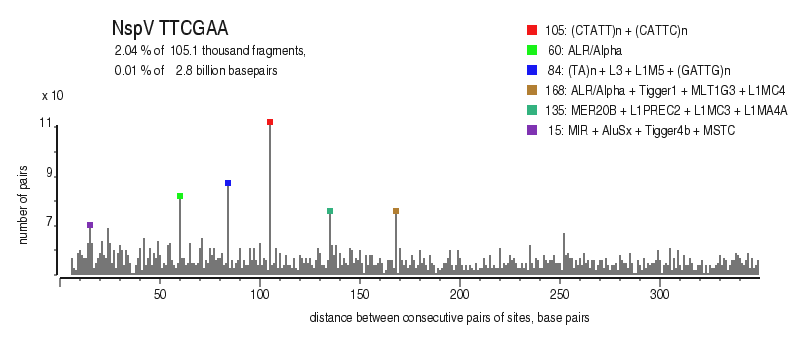
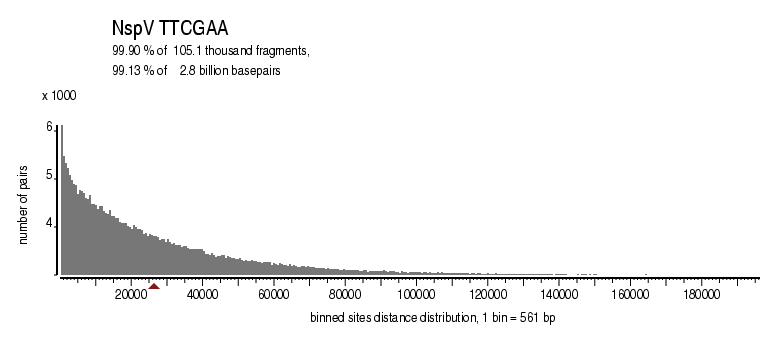
| Enzyme: | NspV | Longest uncut segments |
| Specificity: | TTCGAA | Repeats in uncut segments |
| Number of sites: | 105070 | Genes in uncut segments |
| Mean distance between sites: | 27232 base pairs | |
| Standard deviation: | 28088 base pairs | |
| Site density | 36.7 per megabase | Help |
| Distribution of closely spaced sites |
|---|
 |
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 516074 | chr15 | NT_037852.6 | 1398403-1914477 | 1.92 % in | 46 repeats | 1.93 % in | 1 genes |
| 2 | 451729 | chr6 | NT_167244.1 | 2338469-2790198 | 5.52 % in | 99 repeats | 3.00 % in | 2 genes |
| 3 | 437031 | chrY | NT_011875.12 | 8354113-8791144 | 80.34 % in | 221 repeats | 0.45 % in | 3 genes |
| 4 | 383109 | chr1 | NT_004350.19 | 1824554-2207663 | 16.75 % in | 280 repeats | 67.48 % in | 10 genes |
| 5 | 348512 | chrY | NT_011875.12 | 337594-686106 | 65.59 % in | 700 repeats | 11.53 % in | 5 genes |
| 6 | 342508 | chr4 | NT_016354.19 | 80307263-80649771 | 58.26 % in | 552 repeats | 0.00 % in | 0 genes |
| 7 | 337550 | chr6 | NT_167244.1 | 1995519-2333069 | 16.08 % in | 196 repeats | 8.64 % in | 6 genes |
| 8 | 330722 | chr13 | NT_009952.14 | 11512451-11843173 | 58.35 % in | 764 repeats | 21.54 % in | 2 genes |
| 9 | 327049 | chr6 | NT_167244.1 | 3039544-3366593 | 17.76 % in | 290 repeats | 0.00 % in | 0 genes |
| 10 | 326497 | chr11 | NT_009237.18 | 49612773-49939270 | 53.43 % in | 462 repeats | 0.00 % in | 0 genes |
| 11 | 310036 | chrY | NT_011896.9 | 5824992-6135028 | 33.19 % in | 388 repeats | 0.00 % in | 0 genes |
| 12 | 303493 | chr6 | NT_007299.13 | 224644-528137 | 51.97 % in | 283 repeats | 0.00 % in | 0 genes |
| 13 | 303094 | chr5 | NT_006713.15 | 7018875-7321969 | 57.25 % in | 680 repeats | 0.00 % in | 0 genes |
| 14 | 302969 | chr7 | NT_007933.15 | 47595102-47898071 | 59.80 % in | 455 repeats | 0.00 % in | 0 genes |
| 15 | 300085 | chr5 | NT_006713.15 | 9410338-9710423 | 34.62 % in | 466 repeats | 0.00 % in | 0 genes |
| 16 | 294474 | chr7 | NT_007933.15 | 56711772-57006246 | 60.14 % in | 440 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 516074 | chr15 | NT_037852.6 | 1398403-1914477 | 46 | 27 | AT_rich (5) | L2a (4) | (TA)n (3) | |
| 2 | 451729 | chr6 | NT_167244.1 | 2338469-2790198 | 99 | 53 | AluY (7) | AluSx (5) | AluJb (5) | |
| 3 | 437031 | chrY | NT_011875.12 | 8354113-8791144 | 221 | 87 | AT_rich (20) | LTR12B (17) | AluY (11) | |
| 4 | 383109 | chr1 | NT_004350.19 | 1824554-2207663 | 280 | 99 | MIRb (17) | AluSx (17) | MIR (13) | |
| 5 | 348512 | chrY | NT_011875.12 | 337594-686106 | 700 | 201 | AluSx (54) | AluJo (48) | AT_rich (26) | |
| 6 | 342508 | chr4 | NT_016354.19 | 80307263-80649771 | 552 | 208 | AT_rich (38) | MIRb (19) | L2a (18) | |
| 7 | 337550 | chr6 | NT_167244.1 | 1995519-2333069 | 196 | 73 | AluSx (24) | MIR (10) | AluSp (8) | |
| 8 | 330722 | chr13 | NT_009952.14 | 11512451-11843173 | 764 | 202 | AluSx (88) | AT_rich (31) | AluY (25) | |
| 9 | 327049 | chr6 | NT_167244.1 | 3039544-3366593 | 290 | 67 | AluSx (45) | AluY (18) | AluJo (16) | |
| 10 | 326497 | chr11 | NT_009237.18 | 49612773-49939270 | 462 | 157 | AT_rich (42) | MIRb (24) | L2a (23) | |
| 11 | 310036 | chrY | NT_011896.9 | 5824992-6135028 | 388 | 111 | SST1 (60) | (CA)n (30) | AT_rich (25) | |
| 12 | 303493 | chr6 | NT_007299.13 | 224644-528137 | 283 | 115 | AT_rich (22) | ALR/Alpha (16) | MIR (13) | |
| 13 | 303094 | chr5 | NT_006713.15 | 7018875-7321969 | 680 | 174 | AluSx (75) | AluJb (38) | AluSq (33) | |
| 14 | 302969 | chr7 | NT_007933.15 | 47595102-47898071 | 455 | 167 | AT_rich (34) | L2c (15) | MIRb (13) | |
| 15 | 300085 | chr5 | NT_006713.15 | 9410338-9710423 | 466 | 135 | AT_rich (40) | MIRb (31) | L2a (27) | |
| 16 | 294474 | chr7 | NT_007933.15 | 56711772-57006246 | 440 | 162 | AT_rich (42) | (TA)n (16) | AluSx (13) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 516074 | chr15 | NT_037852.6 | 1398403-1914477 | LOC100418897 | |
| 2 | 451729 | chr6 | NT_167244.1 | 2338469-2790198 | HCG22 | HLA_complex_group_22 |
| MICB | MHC_class_I_polypeptide-related_sequence_B_precursor | |||||
| 3 | 437031 | chrY | NT_011875.12 | 8354113-8791144 | ZNF886P | |
| ZNF884P | ||||||
| ZNF885P | ||||||
| 4 | 383109 | chr1 | NT_004350.19 | 1824554-2207663 | PLCH2 | 1-phosphatidylinositol-4,5-bisphosphate_phosphodiesterase_eta-2 |
| PANK4 | pantothenate_kinase_4 | |||||
| HES5 | transcription_factor_HES-5 | |||||
| LOC115110 | hypothetical_LOC115110 | |||||
| LOC100133445 | hypothetical_LOC100133445 | |||||
| TNFRSF14 | tumor_necrosis_factor_receptor_superfamily_member_14_precursor | |||||
| LOC100506589 | hypothetical_LOC100506589 | |||||
| C1orf93 | hypothetical_protein_LOC127281_isoform_b | |||||
| MMEL1 | membrane_metallo-endopeptidase-like_1 | |||||
| TTC34 | tetratricopeptide_repeat_protein_34 | |||||
| 5 | 348512 | chrY | NT_011875.12 | 337594-686106 | RPS24P1 | |
| ARSFP1 | ||||||
| FAM8A8P | ||||||
| ARSEP1 | ||||||
| ARSDP1 | ||||||
| 7 | 337550 | chr6 | NT_167244.1 | 1995519-2333069 | MDC1 | mediator_of_DNA_damage_checkpoint_protein_1 |
| LOC100294090 | hypothetical_LOC100294090,_transcript_variant_1 | |||||
| FLOT1 | flotillin-1 | |||||
| DDR1 | epithelial_discoidin_domain-containing_receptor_1_isoform_DDR1c | |||||
| MUC21 | mucin-21_precursor | |||||
| LOC100507702 | hypothetical_protein_LOC100507702 | |||||
| 8 | 330722 | chr13 | NT_009952.14 | 11512451-11843173 | RPL7AP61 | |
| FTLP8 | importin-5 | |||||