| Enzyme: | FspI | Longest uncut segments |
| Specificity: | TGCGCA | Repeats in uncut segments |
| Number of sites: | 87588 | Genes in uncut segments |
| Mean distance between sites: | 32668 base pairs | |
| Standard deviation: | 40144 base pairs | |
| Site density | 30.6 per megabase | Help |
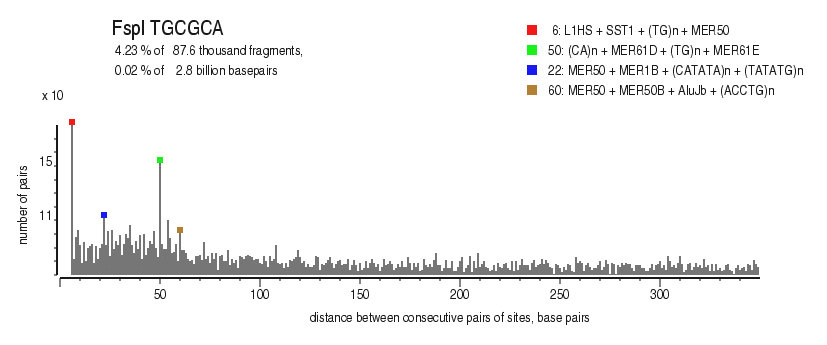
| Distribution of closely spaced sites |
|---|
 |
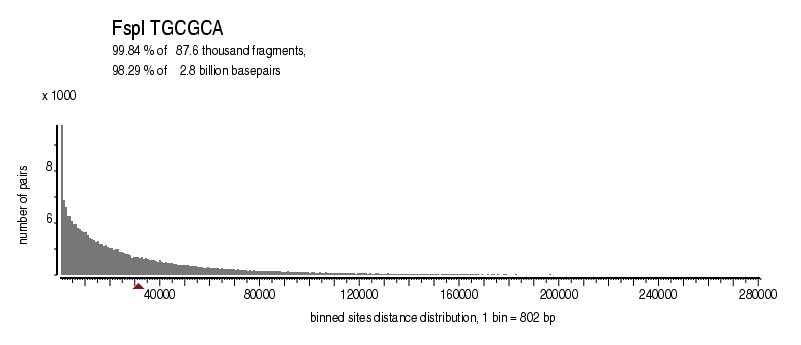
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 745754 | chr4 | NT_016354.19 | 29518364-30264118 | 56.36 % in | 1105 repeats | 2.57 % in | 1 genes |
| 2 | 649184 | chr3 | NT_005612.16 | 41598983-42248167 | 58.93 % in | 1100 repeats | 10.55 % in | 1 genes |
| 3 | 549436 | chr2 | NT_005403.17 | 33286709-33836145 | 49.15 % in | 916 repeats | 64.78 % in | 2 genes |
| 4 | 539256 | chr15 | NT_037852.6 | 1388646-1927902 | 4.12 % in | 87 repeats | 1.85 % in | 1 genes |
| 5 | 488569 | chr1 | NT_032977.9 | 44439688-44928257 | 50.37 % in | 774 repeats | 96.97 % in | 3 genes |
| 6 | 463660 | chr6 | NT_167244.1 | 2324312-2787972 | 6.89 % in | 129 repeats | 2.82 % in | 2 genes |
| 7 | 460997 | chr8 | NT_167187.1 | 31165902-31626899 | 91.94 % in | 237 repeats | 0.00 % in | 0 genes |
| 8 | 455302 | chr6 | NT_025741.15 | 6172139-6627441 | 37.94 % in | 729 repeats | 100.00 % in | 1 genes |
| 9 | 442565 | chrX | NT_011786.16 | 4271809-4714374 | 51.87 % in | 608 repeats | 0.00 % in | 0 genes |
| 10 | 439528 | chr7 | NT_007933.15 | 20914768-21354296 | 40.09 % in | 675 repeats | 0.00 % in | 0 genes |
| 11 | 430362 | chrY | NT_011875.12 | 4350122-4780484 | 64.67 % in | 791 repeats | 0.00 % in | 0 genes |
| 12 | 429474 | chr21 | NT_011512.11 | 15390326-15819800 | 51.47 % in | 724 repeats | 0.00 % in | 0 genes |
| 13 | 428940 | chr8 | NT_008046.16 | 36101428-36530368 | 50.45 % in | 738 repeats | 0.00 % in | 0 genes |
| 14 | 412278 | chr13 | NT_009952.14 | 1006311-1418589 | 50.31 % in | 703 repeats | 0.00 % in | 0 genes |
| 15 | 410452 | chr2 | NT_005403.17 | 23830494-24240946 | 38.62 % in | 662 repeats | 0.00 % in | 0 genes |
| 16 | 407599 | chr14 | NT_026437.12 | 40930959-41338558 | 51.33 % in | 643 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 745754 | chr4 | NT_016354.19 | 29518364-30264118 | 1105 | 265 | AT_rich (114) | L2a (52) | MIRb (46) | |
| 2 | 649184 | chr3 | NT_005612.16 | 41598983-42248167 | 1100 | 269 | L2c (54) | MIRb (47) | L2a (45) | |
| 3 | 549436 | chr2 | NT_005403.17 | 33286709-33836145 | 916 | 236 | AT_rich (87) | L2a (44) | MIRb (41) | |
| 4 | 539256 | chr15 | NT_037852.6 | 1388646-1927902 | 87 | 46 | AT_rich (10) | L1MDa (6) | L2a (5) | |
| 5 | 488569 | chr1 | NT_032977.9 | 44439688-44928257 | 774 | 210 | AT_rich (85) | MIRb (45) | MIR (38) | |
| 6 | 463660 | chr6 | NT_167244.1 | 2324312-2787972 | 129 | 59 | AluSx (8) | AluJo (8) | L1MC4a (7) | |
| 7 | 460997 | chr8 | NT_167187.1 | 31165902-31626899 | 237 | 75 | ALR/Alpha (51) | MSTA (21) | MSTA-int (15) | |
| 8 | 455302 | chr6 | NT_025741.15 | 6172139-6627441 | 729 | 179 | AT_rich (104) | L2a (37) | MIRb (34) | |
| 9 | 442565 | chrX | NT_011786.16 | 4271809-4714374 | 608 | 187 | AluSx (31) | AluY (29) | MIR (22) | |
| 10 | 439528 | chr7 | NT_007933.15 | 20914768-21354296 | 675 | 198 | AT_rich (89) | L2a (32) | MIR (26) | |
| 11 | 430362 | chrY | NT_011875.12 | 4350122-4780484 | 791 | 190 | AT_rich (62) | (TA)n (32) | AluJb (31) | |
| 12 | 429474 | chr21 | NT_011512.11 | 15390326-15819800 | 724 | 218 | AT_rich (43) | AluSx (25) | MIR (24) | |
| 13 | 428940 | chr8 | NT_008046.16 | 36101428-36530368 | 738 | 192 | AT_rich (50) | AluSx (40) | MIRb (39) | |
| 14 | 412278 | chr13 | NT_009952.14 | 1006311-1418589 | 703 | 213 | AT_rich (83) | (TA)n (28) | L2a (26) | |
| 15 | 410452 | chr2 | NT_005403.17 | 23830494-24240946 | 662 | 193 | MIRb (36) | AT_rich (33) | MIR (29) | |
| 16 | 407599 | chr14 | NT_026437.12 | 40930959-41338558 | 643 | 189 | AT_rich (42) | MIRb (35) | MIR (32) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 745754 | chr4 | NT_016354.19 | 29518364-30264118 | CXXC4 | CXXC-type_zinc_finger_protein_4 |
| 2 | 649184 | chr3 | NT_005612.16 | 41598983-42248167 | LOC100420050 | serine/threonine-protein_phosphatase_2A_regulatory_subunit_B''_subunit_alpha_isoform_3 |
| 3 | 549436 | chr2 | NT_005403.17 | 33286709-33836145 | PDE1A | calcium/calmodulin-dependent_3',5'-cyclic_nucleotide_phosphodiesterase_1A_isoform_2 |
| DNAJC10 | dnaJ_homolog_subfamily_C_member_10_precursor | |||||
| 4 | 539256 | chr15 | NT_037852.6 | 1388646-1927902 | LOC100418897 | |
| 5 | 488569 | chr1 | NT_032977.9 | 44439688-44928257 | LRRIQ3 | leucine-rich_repeat_and_IQ_domain-containing_protein_3 |
| FPGT | fucose-1-phosphate_guanylyltransferase | |||||
| TNNI3K | serine/threonine-protein_kinase_TNNI3K_isoform_b | |||||
| 6 | 463660 | chr6 | NT_167244.1 | 2324312-2787972 | HCG22 | HLA_complex_group_22 |
| MICB | MHC_class_I_polypeptide-related_sequence_B_precursor | |||||
| 8 | 455302 | chr6 | NT_025741.15 | 6172139-6627441 | LOC100418924 | glutamate_receptor,_ionotropic_kainate_2_isoform_2_precursor |