| Enzyme: | Sbo13I | Longest uncut segments |
| Specificity: | TCGCGA | Repeats in uncut segments |
| Number of sites: | 14417 | Genes in uncut segments |
| Mean distance between sites: | 198470 base pairs | |
| Standard deviation: | 255722 base pairs | |
| Site density | 5.0 per megabase | Help |
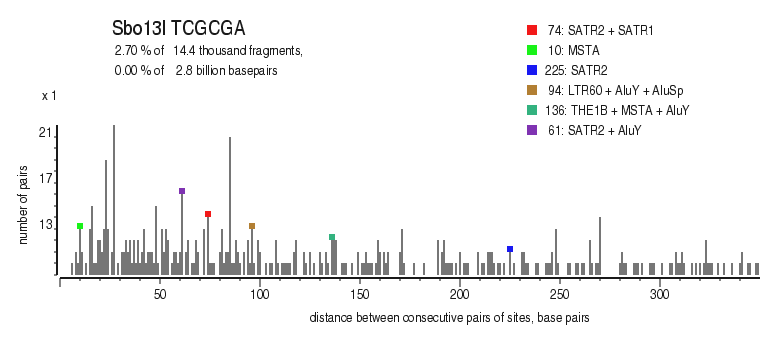
| Distribution of closely spaced sites |
|---|
 |
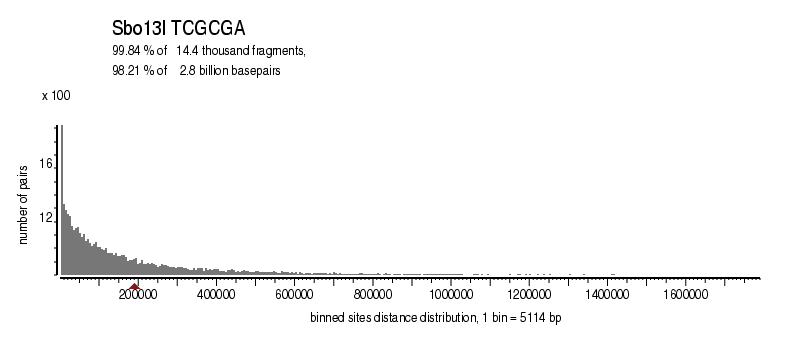
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 2964298 | chr5 | NT_006576.16 | 10980963-13945261 | 47.33 % in | 4549 repeats | 47.18 % in | 5 genes |
| 2 | 2820313 | chrY | NT_011875.12 | 3072843-5893156 | 59.86 % in | 4674 repeats | 4.27 % in | 9 genes |
| 3 | 2432625 | chr4 | NT_022778.16 | 5929802-8362427 | 49.79 % in | 3654 repeats | 19.23 % in | 7 genes |
| 4 | 2388355 | chr2 | NT_022184.15 | 1-2388356 | 46.44 % in | 3739 repeats | 2.18 % in | 4 genes |
| 5 | 2379534 | chr4 | NT_016354.19 | 59050621-61430155 | 52.70 % in | 3518 repeats | 2.41 % in | 7 genes |
| 6 | 2344814 | chr18 | NT_010966.14 | 20344513-22689327 | 44.47 % in | 3726 repeats | 23.47 % in | 6 genes |
| 7 | 2323802 | chr3 | NT_005612.16 | 1693438-4017240 | 51.88 % in | 3389 repeats | 42.16 % in | 10 genes |
| 8 | 2302326 | chr4 | NT_016354.19 | 103276745-105579071 | 52.11 % in | 3750 repeats | 7.97 % in | 2 genes |
| 9 | 2224614 | chr4 | NT_016354.19 | 16996845-19221459 | 49.98 % in | 3443 repeats | 0.00 % in | 0 genes |
| 10 | 2165509 | chr3 | NT_022517.18 | 651313-2816822 | 47.14 % in | 3539 repeats | 0.00 % in | 0 genes |
| 11 | 2145521 | chr11 | NT_009237.18 | 20569033-22714554 | 52.53 % in | 3784 repeats | 0.00 % in | 0 genes |
| 12 | 2089323 | chr2 | NT_005403.17 | 48014624-50103947 | 47.08 % in | 3668 repeats | 0.00 % in | 0 genes |
| 13 | 2080223 | chr2 | NT_022135.16 | 35934033-38014256 | 46.76 % in | 3132 repeats | 0.00 % in | 0 genes |
| 14 | 2015014 | chr5 | NT_034772.6 | 43857354-45872368 | 49.24 % in | 3455 repeats | 0.00 % in | 0 genes |
| 15 | 2010478 | chr13 | NT_024524.14 | 39939616-41950094 | 46.44 % in | 3334 repeats | 0.00 % in | 0 genes |
| 16 | 1999559 | chr14 | NT_026437.12 | 43116735-45116294 | 52.97 % in | 3403 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 2964298 | chr5 | NT_006576.16 | 10980963-13945261 | 4549 | 536 | AT_rich (419) | L2a (183) | AluSx (143) | |
| 2 | 2820313 | chrY | NT_011875.12 | 3072843-5893156 | 4674 | 528 | AT_rich (358) | AluSx (218) | AluJo (190) | |
| 3 | 2432625 | chr4 | NT_022778.16 | 5929802-8362427 | 3654 | 480 | AT_rich (455) | MIR (139) | L2a (133) | |
| 4 | 2388355 | chr2 | NT_022184.15 | 1-2388356 | 3739 | 465 | AT_rich (210) | MIRb (175) | L2c (159) | |
| 5 | 2379534 | chr4 | NT_016354.19 | 59050621-61430155 | 3518 | 484 | AT_rich (525) | L2a (135) | MIR (94) | |
| 6 | 2344814 | chr18 | NT_010966.14 | 20344513-22689327 | 3726 | 494 | AT_rich (296) | MIRb (163) | MIR (157) | |
| 7 | 2323802 | chr3 | NT_005612.16 | 1693438-4017240 | 3389 | 499 | AT_rich (435) | L2a (108) | MIR (100) | |
| 8 | 2302326 | chr4 | NT_016354.19 | 103276745-105579071 | 3750 | 491 | AT_rich (456) | AluSx (126) | L2a (122) | |
| 9 | 2224614 | chr4 | NT_016354.19 | 16996845-19221459 | 3443 | 454 | AT_rich (414) | L2a (179) | MIR (137) | |
| 10 | 2165509 | chr3 | NT_022517.18 | 651313-2816822 | 3539 | 478 | AT_rich (257) | MIR (187) | MIRb (182) | |
| 11 | 2145521 | chr11 | NT_009237.18 | 20569033-22714554 | 3784 | 435 | MIRb (261) | MIR (225) | AT_rich (225) | |
| 12 | 2089323 | chr2 | NT_005403.17 | 48014624-50103947 | 3668 | 442 | AT_rich (266) | AluSx (211) | L2a (138) | |
| 13 | 2080223 | chr2 | NT_022135.16 | 35934033-38014256 | 3132 | 457 | AT_rich (296) | MIRb (126) | MIR (118) | |
| 14 | 2015014 | chr5 | NT_034772.6 | 43857354-45872368 | 3455 | 409 | MIRb (282) | AluSx (186) | MIR (184) | |
| 15 | 2010478 | chr13 | NT_024524.14 | 39939616-41950094 | 3334 | 467 | AT_rich (273) | L2a (155) | MIRb (126) | |
| 16 | 1999559 | chr14 | NT_026437.12 | 43116735-45116294 | 3403 | 442 | AluSx (194) | AT_rich (171) | MIRb (136) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 2964298 | chr5 | NT_006576.16 | 10980963-13945261 | CTNND2 | catenin_delta-2 |
| LOC100420683 | tumor_antigen_gene | |||||
| RPS23P5 | ||||||
| RPL29P13 | ||||||
| DNAH5 | dynein_heavy_chain_5,_axonemal | |||||
| 2 | 2820313 | chrY | NT_011875.12 | 3072843-5893156 | AGKP1 | neuroligin-4,_Y-linked_isoform_2 |
| MED13P1 | ||||||
| CYCSP46 | ||||||
| HDHD1P1 | ||||||
| STSP1 | ||||||
| SURF6P1 | ||||||
| FAM41AY1 | family_with_sequence_similarity_41,_member_A,_Y-linked_1 | |||||
| TUBB1P2 | ||||||
| NCRNA00230B | non-protein_coding_RNA_230B | |||||
| 3 | 2432625 | chr4 | NT_022778.16 | 5929802-8362427 | LOC401134 | hypothetical_LOC401134 |
| LOC100422019 | ||||||
| LOC100507063 | hypothetical_LOC100507063 | |||||
| LOC100144602 | hypothetical_LOC100144602 | |||||
| LOC728048 | ||||||
| MIR1269 | microRNA:hsa-mir-1269 | |||||
| RPS23P3 | ||||||
| 4 | 2388355 | chr2 | NT_022184.15 | 1-2388356 | APOB | apolipoprotein_B-100_precursor |
| LOC100287183 | ||||||
| LOC100507562 | hypothetical_LOC100507562 | |||||
| LOC100130841 | ||||||
| 5 | 2379534 | chr4 | NT_016354.19 | 59050621-61430155 | PABPC4L | polyadenylate-binding_protein_4-like |
| LOC345016 | ||||||
| LOC646272 | ||||||
| RPS2P27 | ||||||
| EEF1A1P35 | ||||||
| LOC100418792 | ||||||
| LOC100507528 | hypothetical_LOC100507528 | |||||
| 6 | 2344814 | chr18 | NT_010966.14 | 20344513-22689327 | KC6 | keratoconus_gene_6 |
| NPM1P1 | ||||||
| LOC100301521 | ||||||
| PIK3C3 | phosphatidylinositol_3-kinase_catalytic_subunit_type_3 | |||||
| RIT2 | GTP-binding_protein_Rit2 | |||||
| SYT4 | synaptotagmin-4 | |||||
| 7 | 2323802 | chr3 | NT_005612.16 | 1693438-4017240 | LOC100287639 | |
| LOC644063 | ||||||
| LOC391556 | ||||||
| MTRNR2L12 | ||||||
| RPL18AP8 | ||||||
| LOC100131442 | ||||||
| LOC100129736 | ||||||
| EPHA6 | ephrin_type-A_receptor_6_isoform_b | |||||
| ARL6 | ADP-ribosylation_factor-like_protein_6 | |||||
| LOC100506362 | hypothetical_LOC100506362 | |||||
| 8 | 2302326 | chr4 | NT_016354.19 | 103276745-105579071 | LOC100506196 | hypothetical_LOC100506196 |
| RPL19P8 | ||||||