| Enzyme: | SnaBI | Longest uncut segments |
| Specificity: | TACGTA | Repeats in uncut segments |
| Number of sites: | 127210 | Genes in uncut segments |
| Mean distance between sites: | 22493 base pairs | |
| Standard deviation: | 25247 base pairs | |
| Site density | 44.5 per megabase | Help |
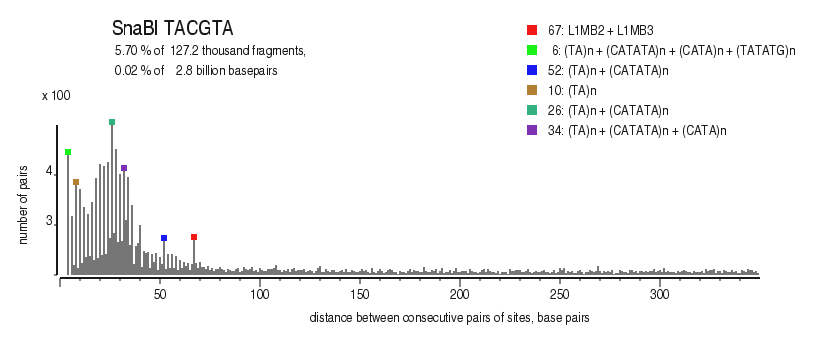
| Distribution of closely spaced sites |
|---|
 |
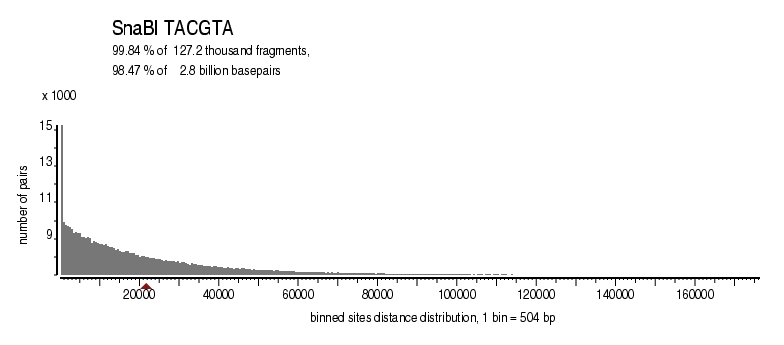
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 523748 | chr15 | NT_037852.6 | 1372768-1896516 | 4.60 % in | 78 repeats | 0.66 % in | 1 genes |
| 2 | 458483 | chr6 | NT_167244.1 | 2324300-2782783 | 5.97 % in | 122 repeats | 1.72 % in | 2 genes |
| 3 | 415199 | chr22 | NT_011520.12 | 22052407-22467606 | 51.37 % in | 1094 repeats | 42.76 % in | 10 genes |
| 4 | 396041 | chr9 | NT_019501.13 | 463155-859196 | 33.60 % in | 513 repeats | 49.85 % in | 3 genes |
| 5 | 359069 | chrX | NT_079573.4 | 1978709-2337778 | 51.22 % in | 679 repeats | 0.00 % in | 0 genes |
| 6 | 355751 | chr19 | NT_011109.16 | 21737882-22093633 | 56.00 % in | 906 repeats | 66.36 % in | 31 genes |
| 7 | 354882 | chr6 | NT_167244.1 | 3078065-3432947 | 15.89 % in | 270 repeats | 21.81 % in | 11 genes |
| 8 | 334449 | chr6 | NT_007592.15 | 37334541-37668990 | 42.74 % in | 698 repeats | 41.00 % in | 5 genes |
| 9 | 318187 | chr11 | NT_009237.18 | 967745-1285932 | 24.60 % in | 311 repeats | 0.00 % in | 0 genes |
| 10 | 311166 | chr19 | NT_011295.11 | 10381920-10693086 | 52.76 % in | 827 repeats | 0.00 % in | 0 genes |
| 11 | 310742 | chr20 | NT_011362.10 | 1239512-1550254 | 51.16 % in | 857 repeats | 0.00 % in | 0 genes |
| 12 | 309932 | chr6 | NT_167244.1 | 3743371-4053303 | 20.42 % in | 219 repeats | 0.00 % in | 0 genes |
| 13 | 305883 | chr6 | NT_167244.1 | 1981239-2287122 | 10.49 % in | 137 repeats | 0.00 % in | 0 genes |
| 14 | 303830 | chr6 | NT_167246.1 | 3157224-3461054 | 15.46 % in | 204 repeats | 0.00 % in | 0 genes |
| 15 | 303493 | chr1 | NT_032977.9 | 802296-1105789 | 56.61 % in | 599 repeats | 0.00 % in | 0 genes |
| 16 | 303235 | chr6 | NT_167246.1 | 2192747-2495982 | 40.99 % in | 523 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 523748 | chr15 | NT_037852.6 | 1372768-1896516 | 78 | 42 | Tigger2 (6) | L1ME3C (6) | L1MDa (6) | |
| 2 | 458483 | chr6 | NT_167244.1 | 2324300-2782783 | 122 | 57 | AluSx (8) | AluJo (8) | L1MC4a (7) | |
| 3 | 415199 | chr22 | NT_011520.12 | 22052407-22467606 | 1094 | 178 | AluSx (140) | MIRb (80) | MIR (53) | |
| 4 | 396041 | chr9 | NT_019501.13 | 463155-859196 | 513 | 158 | AluSx (26) | MIRc (24) | MIRb (22) | |
| 5 | 359069 | chrX | NT_079573.4 | 1978709-2337778 | 679 | 174 | MIR (46) | MIRb (39) | L2c (33) | |
| 6 | 355751 | chr19 | NT_011109.16 | 21737882-22093633 | 906 | 156 | AluSx (140) | AluY (59) | AluJb (45) | |
| 7 | 354882 | chr6 | NT_167244.1 | 3078065-3432947 | 270 | 60 | AluSx (45) | AluSq (16) | AluJo (16) | |
| 8 | 334449 | chr6 | NT_007592.15 | 37334541-37668990 | 698 | 149 | MIR (54) | MIRb (48) | AluSx (48) | |
| 9 | 318187 | chr11 | NT_009237.18 | 967745-1285932 | 311 | 112 | AluSx (30) | L1M5 (12) | GC_rich (11) | |
| 10 | 311166 | chr19 | NT_011295.11 | 10381920-10693086 | 827 | 121 | AluSx (143) | AluJo (77) | AluJb (50) | |
| 11 | 310742 | chr20 | NT_011362.10 | 1239512-1550254 | 857 | 125 | AluSx (91) | MIR (68) | MIRb (62) | |
| 12 | 309932 | chr6 | NT_167244.1 | 3743371-4053303 | 219 | 109 | L2a (18) | AT_rich (15) | AluY (7) | |
| 13 | 305883 | chr6 | NT_167244.1 | 1981239-2287122 | 137 | 58 | AluSx (18) | MIR (7) | AluSg (7) | |
| 14 | 303830 | chr6 | NT_167246.1 | 3157224-3461054 | 204 | 65 | AluSx (32) | AluSq (11) | AluJb (10) | |
| 15 | 303493 | chr1 | NT_032977.9 | 802296-1105789 | 599 | 150 | MIR (79) | MIRb (75) | L2b (46) | |
| 16 | 303235 | chr6 | NT_167246.1 | 2192747-2495982 | 523 | 143 | AluSx (55) | AluJb (29) | AluSq (24) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 523748 | chr15 | NT_037852.6 | 1372768-1896516 | LOC100418897 | |
| 2 | 458483 | chr6 | NT_167244.1 | 2324300-2782783 | HCG22 | HLA_complex_group_22 |
| MICB | MHC_class_I_polypeptide-related_sequence_B_precursor | |||||
| 3 | 415199 | chr22 | NT_011520.12 | 22052407-22467606 | LOC388906 | hypothetical_LOC388906 |
| LOC100134361 | hypothetical_LOC100134361,_transcript_variant_3 | |||||
| NFAM1 | NFAT_activation_molecule_1_precursor | |||||
| LOC100506662 | hypothetical_LOC100506662 | |||||
| RRP7A | ribosomal_RNA-processing_protein_7_homolog_A | |||||
| SERHL2 | serine_hydrolase-like_protein_2 | |||||
| RRP7B | ribosomal_RNA_processing_7_homolog_B_(S._cerevisiae) | |||||
| POLDIP3 | polymerase_delta-interacting_protein_3_isoform_2 | |||||
| RNU12 | RNA,_U12_small_nuclear | |||||
| ATP5L2 | ATP_synthase_subunit_g_2,_mitochondrial | |||||
| 4 | 396041 | chr9 | NT_019501.13 | 463155-859196 | LOC100506554 | hypothetical_LOC100506554 |
| FCN2 | ficolin-2_isoform_b_precursor | |||||
| FCN1 | ficolin-1_precursor | |||||
| 6 | 355751 | chr19 | NT_011109.16 | 21737882-22093633 | FTL | ferritin_light_chain |
| GYS1 | glycogen_[starch]_synthase,_muscle_isoform_2 | |||||
| RUVBL2 | ruvB-like_2 | |||||
| LHB | lutropin_subunit_beta_precursor | |||||
| LOC100287421 | hypothetical_protein_LOC100287421 | |||||
| CGB | choriogonadotropin_subunit_beta_precursor | |||||
| NTF6A | ||||||
| NTF6G | ||||||
| SNAR-G2 | small_ILF3/NF90-associated_RNA_G2 | |||||
| CGB2 | choriogonadotropin_subunit_beta_variant_2 | |||||
| CGB1 | choriogonadotropin_subunit_beta_variant_1 | |||||
| SNAR-G1 | small_ILF3/NF90-associated_RNA_G1 | |||||
| NTF6B | ||||||
| LOC100287453 | ||||||
| CGB5 | chorionic_gonadotropin,_beta_polypeptide_5_precursor | |||||
| LOC100505886 | hypothetical_LOC100505886 | |||||
| CGB8 | chorionic_gonadotropin,_beta_polypeptide_8_precursor | |||||
| LOC100287489 | ||||||
| CGB7 | chorionic_gonadotropin,_beta_polypeptide_7_precursor | |||||
| LOC100287526 | hypothetical_protein_LOC100287526 | |||||
| NTF4 | neurotrophin-4_preproprotein | |||||
| KCNA7 | potassium_voltage-gated_channel_subfamily_A_member_7 | |||||
| SNRNP70 | U1_small_nuclear_ribonucleoprotein_70_kDa | |||||
| LIN7B | protein_lin-7_homolog_B | |||||
| C19orf73 | putative_uncharacterized_protein_C19orf73 | |||||
| PPFIA3 | liprin-alpha-3 | |||||
| HRC | sarcoplasmic_reticulum_histidine-rich_calcium-binding_protein_precursor | |||||
| TRPM4 | transient_receptor_potential_cation_channel_subfamily_M_member_4_isoform_1 | |||||
| SLC6A21P | ||||||
| SUMO1P4 | ||||||
| MIR4324 | microRNA_4324 | |||||
| 7 | 354882 | chr6 | NT_167244.1 | 3078065-3432947 | VARS | valyl-tRNA_synthetase |
| LSM2 | U6_snRNA-associated_Sm-like_protein_LSm2 | |||||
| HSPA1L | heat_shock_70_kDa_protein_1-like | |||||
| HSPA1A | heat_shock_70_kDa_protein_1A/1B | |||||
| HSPA1B | heat_shock_70_kDa_protein_1A/1B | |||||
| C6orf48 | protein_G8 | |||||
| NEU1 | sialidase-1_precursor | |||||
| SLC44A4 | choline_transporter-like_protein_4_isoform_3 | |||||
| EHMT2 | histone-lysine_N-methyltransferase,_H3_lysine-9_specific_3_isoform_b | |||||
| TNXB | tenascin-X_isoform_1_precursor | |||||
| PRRT1 | proline-rich_transmembrane_protein_1 | |||||
| 8 | 334449 | chr6 | NT_007592.15 | 37334541-37668990 | FTSJD2 | cap-specific_mRNA_(nucleoside-2'-O-)-methyltransferase_1 |
| C6orf129 | transmembrane_and_coiled-coil_domain-containing_protein_C6orf129 | |||||
| LOC100505530 | hypothetical_LOC100505530 | |||||
| LOC100505550 | hypothetical_LOC100505550 | |||||
| MDGA1 | MAM_domain-containing_glycosylphosphatidylinositol_anchor_protein_1_precursor | |||||