| Enzyme: | DrdIV | Longest uncut segments |
| Specificity: | TACGAC | Repeats in uncut segments |
| Number of sites: | 86033 | Genes in uncut segments |
| Mean distance between sites: | 33258 base pairs | |
| Standard deviation: | 34457 base pairs | |
| Site density | 30.1 per megabase | Help |
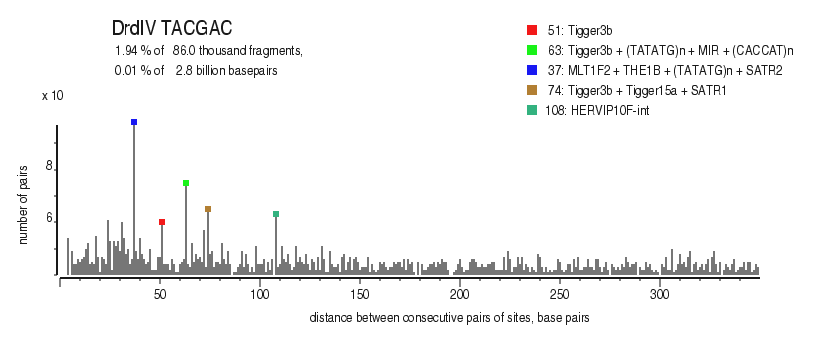
| Distribution of closely spaced sites |
|---|
 |
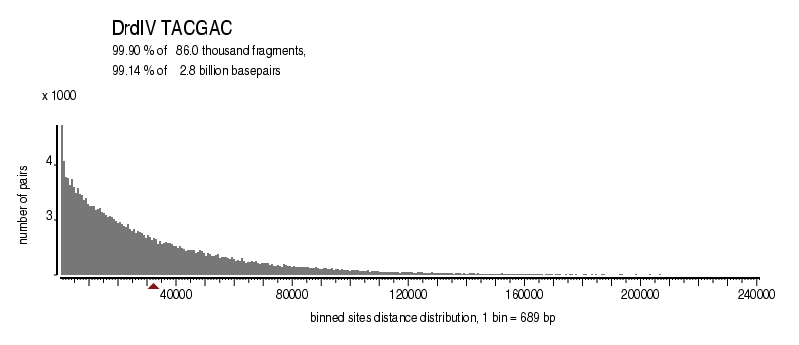
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 638043 | chr15 | NT_037852.6 | 1267796-1905839 | 13.57 % in | 368 repeats | 2.67 % in | 2 genes |
| 2 | 602813 | chr6 | NT_167244.1 | 2281605-2884418 | 14.38 % in | 343 repeats | 8.32 % in | 11 genes |
| 3 | 458601 | chrY | NT_011878.9 | 1-458602 | 35.58 % in | 533 repeats | 26.62 % in | 22 genes |
| 4 | 392877 | chr3 | NT_022517.18 | 10529194-10922071 | 46.46 % in | 841 repeats | 32.31 % in | 2 genes |
| 5 | 385701 | chr12 | NT_029419.12 | 272497-658198 | 92.25 % in | 228 repeats | 2.76 % in | 1 genes |
| 6 | 372929 | chr17 | NT_010799.15 | 2268020-2640949 | 58.61 % in | 998 repeats | 58.52 % in | 9 genes |
| 7 | 372805 | chr11 | NT_167190.1 | 3806761-4179566 | 65.12 % in | 533 repeats | 64.29 % in | 7 genes |
| 8 | 361749 | chr1 | NT_004487.19 | 3369375-3731124 | 48.92 % in | 644 repeats | 17.18 % in | 11 genes |
| 9 | 360979 | chr6 | NT_167249.1 | 2043520-2404499 | 23.96 % in | 364 repeats | 0.00 % in | 0 genes |
| 10 | 358323 | chrX | NT_011669.17 | 1744075-2102398 | 76.72 % in | 527 repeats | 0.00 % in | 0 genes |
| 11 | 357937 | chrX | NT_011669.17 | 8028808-8386745 | 76.85 % in | 666 repeats | 0.00 % in | 0 genes |
| 12 | 353563 | chr9 | NT_008413.18 | 7469738-7823301 | 53.96 % in | 584 repeats | 0.00 % in | 0 genes |
| 13 | 325193 | chr1 | NT_004610.19 | 13380588-13705781 | 59.55 % in | 961 repeats | 0.00 % in | 0 genes |
| 14 | 315708 | chr1 | NT_032977.9 | 90462165-90777873 | 32.48 % in | 451 repeats | 0.00 % in | 0 genes |
| 15 | 314807 | chr6 | NT_167244.1 | 1951696-2266503 | 9.79 % in | 152 repeats | 0.00 % in | 0 genes |
| 16 | 313043 | chr11 | NT_009237.18 | 50312025-50625068 | 97.98 % in | 91 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 638043 | chr15 | NT_037852.6 | 1267796-1905839 | 368 | 111 | AluSx (24) | AT_rich (13) | AluJb (13) | |
| 2 | 602813 | chr6 | NT_167244.1 | 2281605-2884418 | 343 | 108 | AluSx (41) | AluJo (20) | AluJb (16) | |
| 3 | 458601 | chrY | NT_011878.9 | 1-458602 | 533 | 110 | AT_rich (47) | (TG)n (45) | SST1 (44) | |
| 4 | 392877 | chr3 | NT_022517.18 | 10529194-10922071 | 841 | 175 | MIRb (107) | MIR (80) | L2b (57) | |
| 5 | 385701 | chr12 | NT_029419.12 | 272497-658198 | 228 | 78 | ALR/Alpha (45) | AluY (11) | AluSg (11) | |
| 6 | 372929 | chr17 | NT_010799.15 | 2268020-2640949 | 998 | 159 | AluSx (166) | AluJb (85) | AluY (66) | |
| 7 | 372805 | chr11 | NT_167190.1 | 3806761-4179566 | 533 | 142 | MIR (31) | MIRb (21) | AT_rich (21) | |
| 8 | 361749 | chr1 | NT_004487.19 | 3369375-3731124 | 644 | 195 | L2c (47) | AluSx (32) | MIR (27) | |
| 9 | 360979 | chr6 | NT_167249.1 | 2043520-2404499 | 364 | 111 | AluSx (41) | AluJb (25) | AluJo (15) | |
| 10 | 358323 | chrX | NT_011669.17 | 1744075-2102398 | 527 | 138 | L2a (23) | AluJo (22) | L2c (20) | |
| 11 | 357937 | chrX | NT_011669.17 | 8028808-8386745 | 666 | 160 | AluSx (59) | AT_rich (23) | AluJb (19) | |
| 12 | 353563 | chr9 | NT_008413.18 | 7469738-7823301 | 584 | 181 | MIRb (50) | L2c (30) | MIRc (22) | |
| 13 | 325193 | chr1 | NT_004610.19 | 13380588-13705781 | 961 | 136 | AluSx (135) | AluJb (69) | MIRb (60) | |
| 14 | 315708 | chr1 | NT_032977.9 | 90462165-90777873 | 451 | 118 | MIRb (38) | L2c (31) | L2a (30) | |
| 15 | 314807 | chr6 | NT_167244.1 | 1951696-2266503 | 152 | 50 | AluSx (23) | AluJb (11) | AluY (8) | |
| 16 | 313043 | chr11 | NT_009237.18 | 50312025-50625068 | 91 | 31 | ALR/Alpha (34) | L1PA4 (7) | MER11C (6) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 638043 | chr15 | NT_037852.6 | 1267796-1905839 | LOC727914 | |
| LOC100418897 | ||||||
| 2 | 602813 | chr6 | NT_167244.1 | 2281605-2884418 | LOC100507702 | hypothetical_protein_LOC100507702 |
| HCG22 | HLA_complex_group_22 | |||||
| MICB | MHC_class_I_polypeptide-related_sequence_B_precursor | |||||
| PPIAP9 | ||||||
| RPL15P4 | ||||||
| MCCD1 | mitochondrial_coiled-coil_domain_protein_1_precursor | |||||
| SNORD117 | small_nucleolar_RNA,_C/D_box_117 | |||||
| TNF | tumor_necrosis_factor | |||||
| LTB | lymphotoxin-beta_isoform_b | |||||
| LST1 | leukocyte-specific_transcript_1_protein_isoform_5 | |||||
| NCR3 | natural_cytotoxicity_triggering_receptor_3_isoform_c | |||||
| 3 | 458601 | chrY | NT_011878.9 | 1-458602 | FAM197Y5 | testis-specific_Y-encoded_protein_1-like |
| TSPY1 | testis-specific_Y-encoded_protein_1_isoform_TSPY-L | |||||
| FAM197Y4 | testis-specific_Y-encoded_protein_1-like | |||||
| TSPY9 | ||||||
| FAM197Y3 | testis-specific_Y-encoded_protein_1-like | |||||
| TSPY6P | ||||||
| FAM197Y2P | family_with_sequence_similarity_197,_Y-linked,_member_2,_pseudogene | |||||
| TSPY10 | testis-specific_Y-encoded_protein_1-like_isoform_2 | |||||
| FAM197Y1 | hypothetical_protein_LOC100289265 | |||||
| TSPY15P | ||||||
| RBMY3AP | RNA_binding_motif_protein,_Y-linked,_family_3,_member_A_pseudogene | |||||
| TSPY16P | ||||||
| TTTY8 | testis-specific_transcript,_Y-linked_8_(non-protein_coding) | |||||
| TTTY7B | testis-specific_transcript,_Y-linked_7B_(non-protein_coding) | |||||
| TTTY21 | testis-specific_transcript,_Y-linked_21_(non-protein_coding) | |||||
| TTTY2 | testis-specific_transcript,_Y-linked_2_(non-protein_coding) | |||||
| TTTY1 | testis-specific_transcript,_Y-linked_1_(non-protein_coding) | |||||
| TTTY22 | testis-specific_transcript,_Y-linked_22_(non-protein_coding) | |||||
| RBMY2NP | ||||||
| TSPY18P | ||||||
| TSPY13P | ||||||
| TTTY23 | testis-specific_transcript,_Y-linked_23_(non-protein_coding) | |||||
| 4 | 392877 | chr3 | NT_022517.18 | 10529194-10922071 | LOC285370 | hypothetical_LOC285370 |
| SLC6A11 | sodium-_and_chloride-dependent_GABA_transporter_3 | |||||
| 5 | 385701 | chr12 | NT_029419.12 | 272497-658198 | LOC100128817 | |
| 6 | 372929 | chr17 | NT_010799.15 | 2268020-2640949 | TWF1P1 | |
| CRYBA1 | beta-crystallin_A3 | |||||
| NUFIP2 | nuclear_fragile_X_mental_retardation-interacting_protein_2 | |||||
| RPL35AP35 | ||||||
| TAOK1 | serine/threonine-protein_kinase_TAO1 | |||||
| LOC100506461 | hypothetical_LOC100506461 | |||||
| ABHD15 | abhydrolase_domain-containing_protein_15_precursor | |||||
| TP53I13 | tumor_protein_p53-inducible_protein_13_precursor | |||||
| GIT1 | ARF_GTPase-activating_protein_GIT1_isoform_2 | |||||
| 7 | 372805 | chr11 | NT_167190.1 | 3806761-4179566 | LOC100420019 | |
| LOC100101405 | ||||||
| GLYATL2 | glycine_N-acyltransferase-like_protein_2 | |||||
| GLYATL1P2 | ||||||
| LOC100287485 | glycine_N-acyltransferase-like_protein_1 | |||||
| GLYATL1P4 | glycine-N-acyltransferase-like_1_pseudogene_4_isoform_3 | |||||
| GLYATL1P3 | ||||||
| 8 | 361749 | chr1 | NT_004487.19 | 3369375-3731124 | THEM4 | thioesterase_superfamily_member_4 |
| KRT8P28 | ||||||
| S100A10 | protein_S100-A10 | |||||
| NBPF18P | ||||||
| S100A11 | protein_S100-A11 | |||||
| LOC100131107 | putative_UPF0607_protein_ENSP00000383783 | |||||
| TCHHL1 | trichohyalin-like_protein_1 | |||||
| TCHH | trichohyalin | |||||
| HDHD1P2 | ||||||
| RPTN | repetin | |||||
| HRNR | hornerin | |||||