| Enzyme: | XamI | Longest uncut segments |
| Specificity: | GTCGAC | Repeats in uncut segments |
| Number of sites: | 30676 | Genes in uncut segments |
| Mean distance between sites: | 93276 base pairs | |
| Standard deviation: | 101295 base pairs | |
| Site density | 10.7 per megabase | Help |
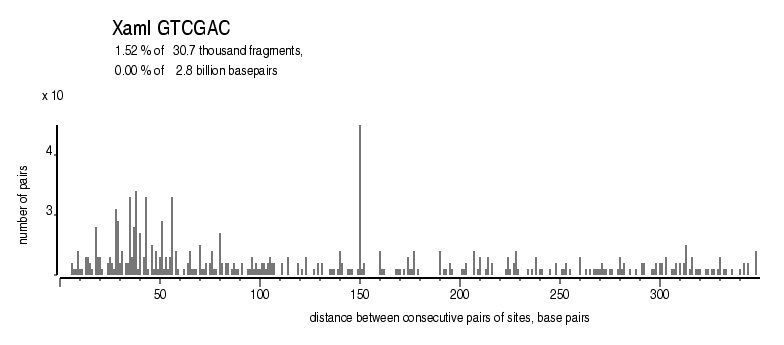
| Distribution of closely spaced sites |
|---|
 |
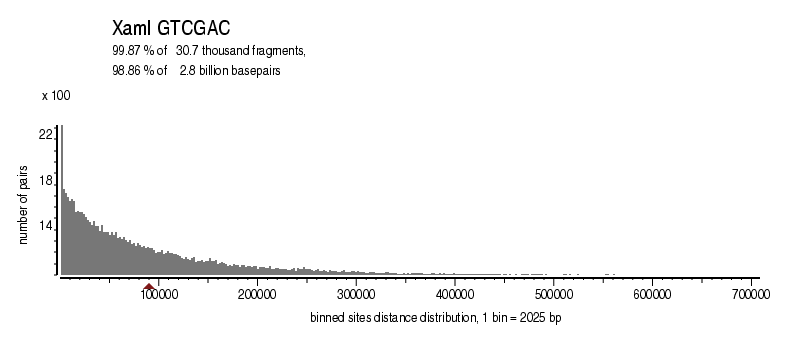
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 1343037 | chr1 | NT_032977.9 | 21443537-22786574 | 61.81 % in | 3520 repeats | 70.86 % in | 13 genes |
| 2 | 1095871 | chrX | NT_167197.1 | 31284761-32380632 | 62.79 % in | 1649 repeats | 0.48 % in | 4 genes |
| 3 | 994289 | chr3 | NT_022517.18 | 5184741-6179030 | 54.28 % in | 1736 repeats | 1.71 % in | 2 genes |
| 4 | 953599 | chr12 | NT_009714.17 | 3484789-4438388 | 52.38 % in | 1479 repeats | 50.10 % in | 20 genes |
| 5 | 931374 | chr11 | NT_033899.8 | 25131799-26063173 | 49.98 % in | 1594 repeats | 39.00 % in | 4 genes |
| 6 | 913850 | chr14 | NT_026437.12 | 26921614-27835464 | 63.19 % in | 1339 repeats | 0.00 % in | 0 genes |
| 7 | 894612 | chr5 | NT_006713.15 | 1393635-2288247 | 48.12 % in | 1334 repeats | 0.18 % in | 2 genes |
| 8 | 885627 | chr9 | NT_008413.18 | 31502686-32388313 | 57.10 % in | 1346 repeats | 1.79 % in | 3 genes |
| 9 | 880188 | chr3 | NT_022459.15 | 19461230-20341418 | 44.56 % in | 1291 repeats | 0.00 % in | 0 genes |
| 10 | 870069 | chr7 | NT_007933.15 | 26466350-27336419 | 51.23 % in | 1329 repeats | 0.00 % in | 0 genes |
| 11 | 852834 | chr8 | NT_167187.1 | 17810991-18663825 | 54.03 % in | 2008 repeats | 0.00 % in | 0 genes |
| 12 | 841025 | chr2 | NT_022184.15 | 10053563-10894588 | 59.31 % in | 1323 repeats | 0.00 % in | 0 genes |
| 13 | 834719 | chr6 | NT_007592.15 | 44034782-44869501 | 46.65 % in | 1506 repeats | 0.00 % in | 0 genes |
| 14 | 823974 | chr5 | NT_034772.6 | 8196414-9020388 | 54.58 % in | 1181 repeats | 0.00 % in | 0 genes |
| 15 | 823485 | chr9 | NT_008470.19 | 34741462-35564947 | 58.87 % in | 1320 repeats | 0.00 % in | 0 genes |
| 16 | 821399 | chr6 | NT_025741.15 | 1-821400 | 51.54 % in | 1288 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 1343037 | chr1 | NT_032977.9 | 21443537-22786574 | 3520 | 380 | AluSx (398) | MIRb (193) | AluJb (153) | |
| 2 | 1095871 | chrX | NT_167197.1 | 31284761-32380632 | 1649 | 334 | AT_rich (132) | (TA)n (38) | MIRb (36) | |
| 3 | 994289 | chr3 | NT_022517.18 | 5184741-6179030 | 1736 | 352 | MIRb (101) | MIR (78) | AT_rich (71) | |
| 4 | 953599 | chr12 | NT_009714.17 | 3484789-4438388 | 1479 | 291 | AT_rich (110) | L2 (53) | MIR (51) | |
| 5 | 931374 | chr11 | NT_033899.8 | 25131799-26063173 | 1594 | 298 | MIRb (109) | AluSx (88) | L2c (78) | |
| 6 | 913850 | chr14 | NT_026437.12 | 26921614-27835464 | 1339 | 305 | AT_rich (126) | L2a (48) | MIRb (39) | |
| 7 | 894612 | chr5 | NT_006713.15 | 1393635-2288247 | 1334 | 290 | AT_rich (162) | MIR (50) | L2a (49) | |
| 8 | 885627 | chr9 | NT_008413.18 | 31502686-32388313 | 1346 | 323 | AT_rich (94) | MIRb (50) | MIR (37) | |
| 9 | 880188 | chr3 | NT_022459.15 | 19461230-20341418 | 1291 | 286 | AT_rich (162) | L2a (47) | (TA)n (46) | |
| 10 | 870069 | chr7 | NT_007933.15 | 26466350-27336419 | 1329 | 290 | AT_rich (153) | MIR (52) | MIRb (42) | |
| 11 | 852834 | chr8 | NT_167187.1 | 17810991-18663825 | 2008 | 284 | AluSx (240) | AluJo (139) | AluJb (119) | |
| 12 | 841025 | chr2 | NT_022184.15 | 10053563-10894588 | 1323 | 294 | MIRb (89) | AluSx (53) | MIR (41) | |
| 13 | 834719 | chr6 | NT_007592.15 | 44034782-44869501 | 1506 | 286 | AluSx (103) | MIRb (89) | MIR (72) | |
| 14 | 823974 | chr5 | NT_034772.6 | 8196414-9020388 | 1181 | 282 | AT_rich (125) | (TA)n (36) | MIR (29) | |
| 15 | 823485 | chr9 | NT_008470.19 | 34741462-35564947 | 1320 | 313 | AT_rich (124) | AluSx (38) | (TA)n (37) | |
| 16 | 821399 | chr6 | NT_025741.15 | 1-821400 | 1288 | 285 | AT_rich (143) | MIR (53) | L2c (40) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 1343037 | chr1 | NT_032977.9 | 21443537-22786574 | C1orf185 | hypothetical_protein_LOC284546 |
| CFLP2 | ||||||
| LOC100128235 | ||||||
| RPS2P8 | RING_finger_protein_11 | |||||
| TTC39A | tetratricopeptide_repeat_protein_39A_isoform_2 | |||||
| LOC100422413 | epidermal_growth_factor_receptor_substrate_15_isoform_B | |||||
| CALR4P | ||||||
| OSBPL9 | oxysterol-binding_protein-related_protein_9_isoform_b | |||||
| MIR761 | microRNA_761 | |||||
| RPS26P14 | ras-related_protein_Rab-3B | |||||
| KTI12 | protein_KTI12_homolog | |||||
| BTF3L4 | transcription_factor_BTF3_homolog_4_isoform_2 | |||||
| ANAPC10P1 | zinc_finger_FYVE_domain-containing_protein_9_isoform_2 | |||||
| 2 | 1095871 | chrX | NT_167197.1 | 31284761-32380632 | LOC646506 | |
| FAM47A | hypothetical_protein_LOC158724 | |||||
| FTH1P14 | ||||||
| LOC392439 | ||||||
| 3 | 994289 | chr3 | NT_022517.18 | 5184741-6179030 | EDEM1 | ER_degradation-enhancing_alpha-mannosidase-like_1 |
| MRPS35P1 | ||||||
| 4 | 953599 | chr12 | NT_009714.17 | 3484789-4438388 | KLRA1 | killer_cell_lectin-like_receptor_subfamily_A,_member_1,_pseudogene |
| MAGOHB | protein_mago_nashi_homolog_2 | |||||
| STYK1 | tyrosine-protein_kinase_STYK1 | |||||
| CSDA | DNA-binding_protein_A_isoform_b | |||||
| LOC100286954 | hypothetical_LOC100506226,_transcript_variant_1 | |||||
| TAS2R7 | taste_receptor_type_2_member_7 | |||||
| TAS2R8 | taste_receptor_type_2_member_8 | |||||
| TAS2R9 | taste_receptor_type_2_member_9 | |||||
| TAS2R10 | taste_receptor_type_2_member_10 | |||||
| LOC100420580 | ||||||
| TAS2R18 | taste_receptor_type_2_member_30 | |||||
| PS5 | ||||||
| TAS2R42 | taste_receptor_type_2_member_42 | |||||
| LOC100420583 | ||||||
| PRB3 | basic_salivary_proline-rich_protein_3_precursor | |||||
| PRB4 | basic_salivary_proline-rich_protein_4_precursor | |||||
| PRB1 | basic_salivary_proline-rich_protein_1_isoform_3_preproprotein | |||||
| PRB2 | basic_salivary_proline-rich_protein_2 | |||||
| LOC100130073 | ||||||
| LOC100129640 | ||||||
| 5 | 931374 | chr11 | NT_033899.8 | 25131799-26063173 | LOC100507165 | hypothetical_LOC100507165 |
| MIR100 | microRNA:hsa-mir-100 | |||||
| - | tRNA-Lys | |||||
| GLULP3 | ||||||
| 7 | 894612 | chr5 | NT_006713.15 | 1393635-2288247 | LOC100128982 | |
| RPS17P11 | ||||||
| 8 | 885627 | chr9 | NT_008413.18 | 31502686-32388313 | LOC100288563 | |
| SLC25A5P8 | ||||||
| ACO1 | cytoplasmic_aconitate_hydratase | |||||