| Enzyme: | BstZ17I | Longest uncut segments |
| Specificity: | GTATAC | Repeats in uncut segments |
| Number of sites: | 424330 | Genes in uncut segments |
| Mean distance between sites: | 6743 base pairs | |
| Standard deviation: | 8247 base pairs | |
| Site density | 148.3 per megabase | Help |
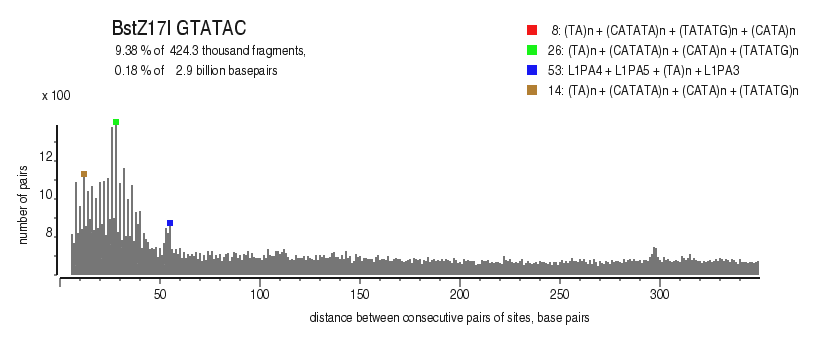
| Distribution of closely spaced sites |
|---|
 |
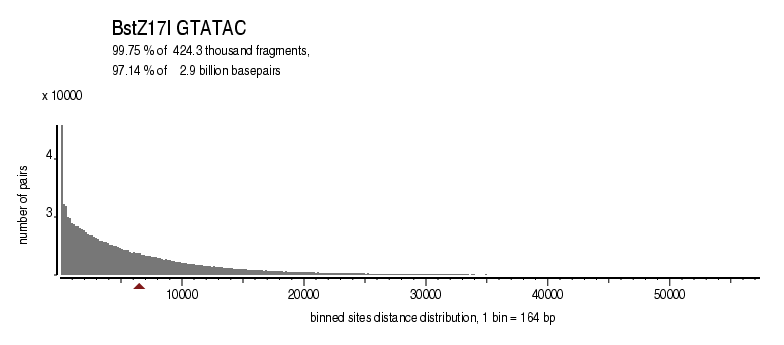
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 496298 | chr15 | NT_037852.6 | 1389323-1885621 | 1.38 % in | 17 repeats | 0.00 % in | 0 genes |
| 2 | 408531 | chr6 | NT_167244.1 | 2357950-2766481 | 0.87 % in | 14 repeats | 0.00 % in | 0 genes |
| 3 | 305998 | chrY | NT_011875.12 | 8416345-8722343 | 82.98 % in | 42 repeats | 0.26 % in | 1 genes |
| 4 | 291266 | chr6 | NT_167244.1 | 3150305-3441571 | 10.71 % in | 155 repeats | 17.96 % in | 6 genes |
| 5 | 268217 | chr8 | NT_023678.16 | 116740-384957 | 99.52 % in | 85 repeats | 0.00 % in | 0 genes |
| 6 | 250600 | chr6 | NT_167244.1 | 2004611-2255211 | 1.91 % in | 23 repeats | 3.11 % in | 3 genes |
| 7 | 245004 | chr19 | NT_011255.14 | 1015795-1260799 | 30.84 % in | 417 repeats | 73.66 % in | 14 genes |
| 8 | 241039 | chr14 | NT_026437.12 | 86690895-86931934 | 37.73 % in | 399 repeats | 92.61 % in | 5 genes |
| 9 | 240221 | chr6 | NT_167244.1 | 2803406-3043627 | 12.80 % in | 156 repeats | 0.00 % in | 0 genes |
| 10 | 228427 | chr9 | NT_008470.19 | 61229631-61458058 | 51.57 % in | 645 repeats | 0.00 % in | 0 genes |
| 11 | 213555 | chr6 | NT_167244.1 | 4388426-4601981 | 2.15 % in | 15 repeats | 0.00 % in | 0 genes |
| 12 | 195418 | chr1 | NT_004350.19 | 1581070-1776488 | 24.39 % in | 214 repeats | 0.00 % in | 0 genes |
| 13 | 191906 | chr6 | NT_167247.1 | 4420255-4612161 | 2.86 % in | 30 repeats | 0.00 % in | 0 genes |
| 14 | 190821 | chr7 | NT_007819.17 | 1493967-1684788 | 37.59 % in | 352 repeats | 0.00 % in | 0 genes |
| 15 | 189162 | chr19 | NT_011295.11 | 9162566-9351728 | 60.71 % in | 576 repeats | 0.00 % in | 0 genes |
| 16 | 188892 | chr6 | NT_167244.1 | 3789514-3978406 | 2.60 % in | 22 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 496298 | chr15 | NT_037852.6 | 1389323-1885621 | 17 | 12 | L1MDa (6) | MIRc (1) | MIRb (1) | |
| 2 | 408531 | chr6 | NT_167244.1 | 2357950-2766481 | 14 | 10 | LTR84b (2) | L4 (2) | AluY (2) | |
| 3 | 305998 | chrY | NT_011875.12 | 8416345-8722343 | 42 | 15 | LTR12B (17) | L1PA16 (6) | L1PA7 (3) | |
| 4 | 291266 | chr6 | NT_167244.1 | 3150305-3441571 | 155 | 49 | AluSx (21) | MIR (9) | AluSq (9) | |
| 5 | 268217 | chr8 | NT_023678.16 | 116740-384957 | 85 | 22 | GSAT (30) | ALR/Alpha (23) | LTR61 (4) | |
| 6 | 250600 | chr6 | NT_167244.1 | 2004611-2255211 | 23 | 16 | AluSx (4) | MIRb (2) | MIR (2) | |
| 7 | 245004 | chr19 | NT_011255.14 | 1015795-1260799 | 417 | 88 | AluSx (54) | GC_rich (30) | AluY (29) | |
| 8 | 241039 | chr14 | NT_026437.12 | 86690895-86931934 | 399 | 90 | AluSx (43) | AluJb (36) | AluY (26) | |
| 9 | 240221 | chr6 | NT_167244.1 | 2803406-3043627 | 156 | 48 | AluSx (22) | AluJo (15) | AluJb (10) | |
| 10 | 228427 | chr9 | NT_008470.19 | 61229631-61458058 | 645 | 127 | MIRb (60) | AluSx (57) | MIR (54) | |
| 11 | 213555 | chr6 | NT_167244.1 | 4388426-4601981 | 15 | 11 | MER57-int (2) | HERVH-int (2) | AluSx (2) | |
| 12 | 195418 | chr1 | NT_004350.19 | 1581070-1776488 | 214 | 72 | AluSx (27) | AluY (16) | AluJo (11) | |
| 13 | 191906 | chr6 | NT_167247.1 | 4420255-4612161 | 30 | 22 | AluSx (3) | MLT1J (2) | MamRep38 (2) | |
| 14 | 190821 | chr7 | NT_007819.17 | 1493967-1684788 | 352 | 85 | AluSx (43) | AluJo (34) | AluJb (19) | |
| 15 | 189162 | chr19 | NT_011295.11 | 9162566-9351728 | 576 | 86 | AluSx (127) | AluJo (52) | AluJb (36) | |
| 16 | 188892 | chr6 | NT_167244.1 | 3789514-3978406 | 22 | 17 | L2a (4) | MLT1H-int (2) | L1M5 (2) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 3 | 305998 | chrY | NT_011875.12 | 8416345-8722343 | ZNF884P | |
| 4 | 291266 | chr6 | NT_167244.1 | 3150305-3441571 | SLC44A4 | choline_transporter-like_protein_4_isoform_3 |
| EHMT2 | histone-lysine_N-methyltransferase,_H3_lysine-9_specific_3_isoform_b | |||||
| TNXB | tenascin-X_isoform_1_precursor | |||||
| PRRT1 | proline-rich_transmembrane_protein_1 | |||||
| LOC100294105 | hypothetical_LOC100294105,_transcript_variant_2 | |||||
| PPT2 | lysosomal_thioesterase_PPT2_isoform_a_precursor | |||||
| 6 | 250600 | chr6 | NT_167244.1 | 2004611-2255211 | LOC100294090 | hypothetical_LOC100294090,_transcript_variant_1 |
| FLOT1 | flotillin-1 | |||||
| DDR1 | epithelial_discoidin_domain-containing_receptor_1_isoform_DDR1c | |||||
| 7 | 245004 | chr19 | NT_011255.14 | 1015795-1260799 | HMHA1 | minor_histocompatibility_protein_HA-1 |
| POLR2E | DNA-directed_RNA_polymerases_I,_II,_and_III_subunit_RPABC1 | |||||
| GPX4 | phospholipid_hydroperoxide_glutathione_peroxidase,_mitochondrial_isoform_C_precursor | |||||
| SBNO2 | protein_strawberry_notch_homolog_2_isoform_2 | |||||
| LOC729119 | ||||||
| STK11 | serine/threonine-protein_kinase_11 | |||||
| C19orf26 | protein_Dos | |||||
| ATP5D | ATP_synthase_subunit_delta,_mitochondrial_precursor | |||||
| MIDN | midnolin | |||||
| C19orf23 | chromosome_19_open_reading_frame_23 | |||||
| CIRBP | cold-inducible_RNA-binding_protein | |||||
| C19orf24 | hypothetical_protein_LOC55009 | |||||
| EFNA2 | ephrin-A2_precursor | |||||
| LOC100287367 | hypothetical_protein_LOC100287367 | |||||
| 8 | 241039 | chr14 | NT_026437.12 | 86690895-86931934 | BTBD6 | BTB/POZ_domain-containing_protein_6 |
| RPS20P33 | phosphofurin_acidic_cluster_sorting_protein_2_isoform_2 | |||||
| LOC647310 | hypothetical_protein_LOC647310 | |||||
| LOC100507437 | hypothetical_LOC100507437,_transcript_variant_1 | |||||
| LOC100128343 | hypothetical_LOC100128343 | |||||