| Enzyme: | NgoMIV | Longest uncut segments |
| Specificity: | GCCGGC | Repeats in uncut segments |
| Number of sites: | 124193 | Genes in uncut segments |
| Mean distance between sites: | 23039 base pairs | |
| Standard deviation: | 44459 base pairs | |
| Site density | 43.4 per megabase | Help |
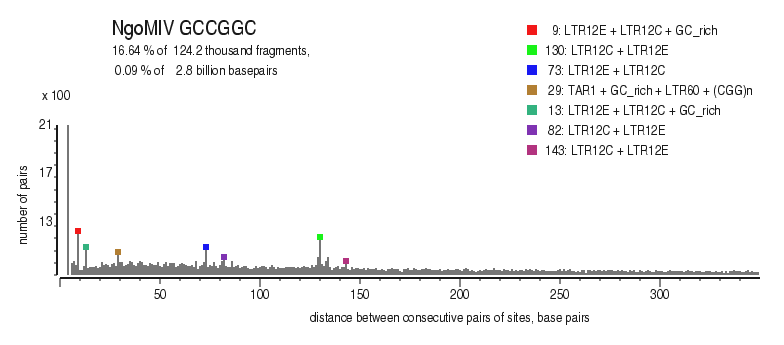
| Distribution of closely spaced sites |
|---|
 |
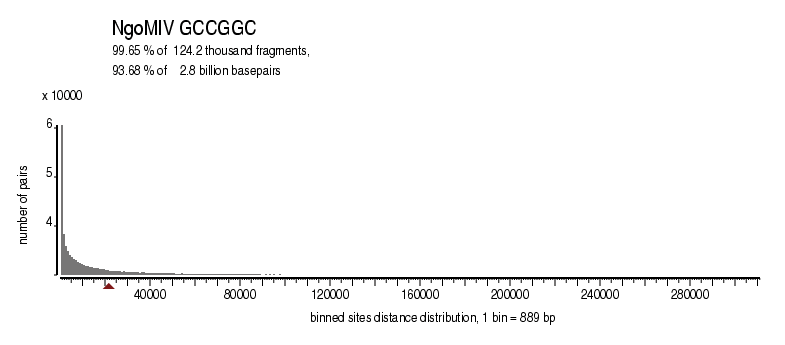
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 1084329 | chr6 | NT_025741.15 | 24448639-25532968 | 58.88 % in | 1613 repeats | 0.05 % in | 2 genes |
| 2 | 920340 | chr2 | NT_022184.15 | 28213087-29133427 | 49.01 % in | 1371 repeats | 18.11 % in | 2 genes |
| 3 | 849911 | chrX | NT_011651.17 | 768557-1618468 | 74.86 % in | 1068 repeats | 10.07 % in | 7 genes |
| 4 | 848302 | chr15 | NT_010194.17 | 8213276-9061578 | 51.60 % in | 1183 repeats | 1.96 % in | 2 genes |
| 5 | 800944 | chr8 | NT_008183.19 | 30057396-30858340 | 52.06 % in | 1106 repeats | 0.00 % in | 0 genes |
| 6 | 777554 | chr14 | NT_026437.12 | 29933558-30711112 | 54.50 % in | 1153 repeats | 0.07 % in | 1 genes |
| 7 | 759809 | chr11 | NT_033899.8 | 310982-1070791 | 59.43 % in | 1092 repeats | 0.00 % in | 0 genes |
| 8 | 754721 | chr12 | NT_029419.12 | 53361332-54116053 | 49.11 % in | 1259 repeats | 13.02 % in | 5 genes |
| 9 | 729489 | chr18 | NT_025028.14 | 16937835-17667324 | 52.06 % in | 1197 repeats | 0.00 % in | 0 genes |
| 10 | 706968 | chr13 | NT_024524.14 | 50880941-51587909 | 51.67 % in | 1157 repeats | 0.00 % in | 0 genes |
| 11 | 675122 | chr2 | NT_022184.15 | 59569798-60244920 | 52.01 % in | 1001 repeats | 0.00 % in | 0 genes |
| 12 | 673298 | chrX | NT_011669.17 | 1-673299 | 94.74 % in | 467 repeats | 0.00 % in | 0 genes |
| 13 | 666577 | chr12 | NT_009714.17 | 26353752-27020329 | 60.57 % in | 971 repeats | 0.00 % in | 0 genes |
| 14 | 651037 | chrX | NT_011669.17 | 13316053-13967090 | 86.96 % in | 720 repeats | 0.00 % in | 0 genes |
| 15 | 645469 | chr11 | NT_009237.18 | 48197832-48843301 | 80.21 % in | 613 repeats | 0.00 % in | 0 genes |
| 16 | 627415 | chr18 | NT_010966.14 | 32332493-32959908 | 49.10 % in | 982 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 1084329 | chr6 | NT_025741.15 | 24448639-25532968 | 1613 | 322 | AT_rich (178) | L1M5 (42) | AluSx (42) | |
| 2 | 920340 | chr2 | NT_022184.15 | 28213087-29133427 | 1371 | 303 | AT_rich (96) | L2a (63) | MIRb (59) | |
| 3 | 849911 | chrX | NT_011651.17 | 768557-1618468 | 1068 | 220 | L2c (53) | MIRb (50) | AT_rich (43) | |
| 4 | 848302 | chr15 | NT_010194.17 | 8213276-9061578 | 1183 | 268 | AT_rich (111) | MIRb (53) | L2a (45) | |
| 5 | 800944 | chr8 | NT_008183.19 | 30057396-30858340 | 1106 | 272 | AT_rich (147) | L2a (44) | MIRb (41) | |
| 6 | 777554 | chr14 | NT_026437.12 | 29933558-30711112 | 1153 | 280 | AT_rich (105) | L2a (48) | L2c (39) | |
| 7 | 759809 | chr11 | NT_033899.8 | 310982-1070791 | 1092 | 294 | AT_rich (84) | MIRb (46) | MIR (43) | |
| 8 | 754721 | chr12 | NT_029419.12 | 53361332-54116053 | 1259 | 280 | AT_rich (113) | MIRb (66) | L2a (53) | |
| 9 | 729489 | chr18 | NT_025028.14 | 16937835-17667324 | 1197 | 285 | AT_rich (140) | AluSx (43) | L2a (37) | |
| 10 | 706968 | chr13 | NT_024524.14 | 50880941-51587909 | 1157 | 298 | AT_rich (141) | L2a (39) | AluSx (36) | |
| 11 | 675122 | chr2 | NT_022184.15 | 59569798-60244920 | 1001 | 280 | AT_rich (74) | MIRb (51) | MIR (46) | |
| 12 | 673298 | chrX | NT_011669.17 | 1-673299 | 467 | 137 | ALR/Alpha (36) | AT_rich (18) | L1M2 (16) | |
| 13 | 666577 | chr12 | NT_009714.17 | 26353752-27020329 | 971 | 259 | AT_rich (84) | L2a (34) | MIRb (33) | |
| 14 | 651037 | chrX | NT_011669.17 | 13316053-13967090 | 720 | 203 | L1PB1 (31) | L1PA7 (26) | L1M1 (21) | |
| 15 | 645469 | chr11 | NT_009237.18 | 48197832-48843301 | 613 | 160 | ALR/Alpha (28) | L1PA4 (26) | L1M5 (25) | |
| 16 | 627415 | chr18 | NT_010966.14 | 32332493-32959908 | 982 | 265 | AT_rich (79) | MIRb (36) | L2a (33) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 1084329 | chr6 | NT_025741.15 | 24448639-25532968 | MIR3144 | microRNA_3144 |
| COX6A1P3 | ||||||
| 2 | 920340 | chr2 | NT_022184.15 | 28213087-29133427 | RPL7P13 | |
| NRXN1 | neurexin-1-beta_isoform_beta_precursor | |||||
| 3 | 849911 | chrX | NT_011651.17 | 768557-1618468 | CYSLTR1 | cysteinyl_leukotriene_receptor_1 |
| ZCCHC5 | zinc_finger_CCHC_domain-containing_protein_5 | |||||
| LOC100421601 | ||||||
| LPAR4 | lysophosphatidic_acid_receptor_4 | |||||
| RPL7P54 | ||||||
| MIR4328 | microRNA_4328 | |||||
| P2RY10 | putative_P2Y_purinoceptor_10 | |||||
| 4 | 848302 | chr15 | NT_010194.17 | 8213276-9061578 | RPS15P8 | |
| TMCO5A | transmembrane_and_coiled-coil_domain-containing_protein_5A | |||||
| 6 | 777554 | chr14 | NT_026437.12 | 29933558-30711112 | RPL18P1 | |
| 8 | 754721 | chr12 | NT_029419.12 | 53361332-54116053 | C12orf12 | hypothetical_protein_LOC196477 |
| EPYC | epiphycan_precursor | |||||
| KERA | keratocan_precursor | |||||
| LUM | lumican_precursor | |||||
| DCN | decorin_isoform_e_precursor | |||||