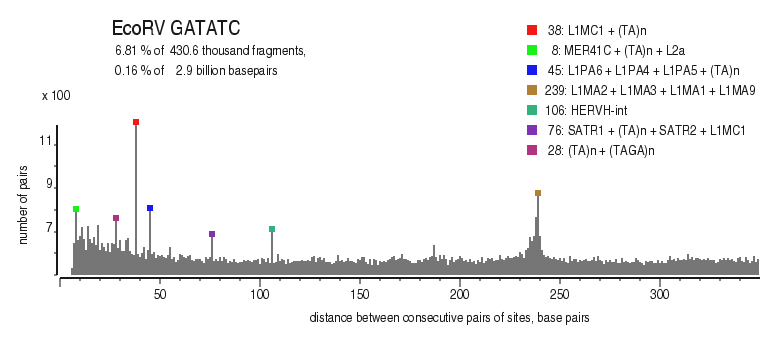
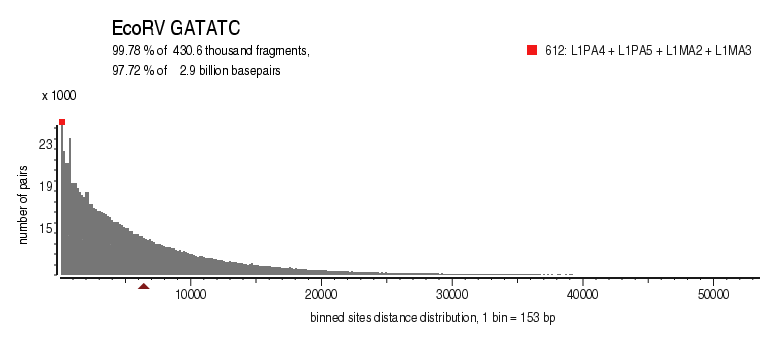
| Enzyme: | EcoRV | Longest uncut segments |
| Specificity: | GATATC | Repeats in uncut segments |
| Number of sites: | 430614 | Genes in uncut segments |
| Mean distance between sites: | 6644 base pairs | |
| Standard deviation: | 7674 base pairs | |
| Site density | 150.5 per megabase | Help |
| Distribution of closely spaced sites |
|---|
 |
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 490064 | chr15 | NT_037852.6 | 1397638-1887702 | 0.17 % in | 6 repeats | 0.00 % in | 0 genes |
| 2 | 430842 | chr6 | NT_167244.1 | 2334074-2764916 | 3.51 % in | 66 repeats | 1.32 % in | 1 genes |
| 3 | 271609 | chr6 | NT_167244.1 | 2003105-2274714 | 5.49 % in | 63 repeats | 5.70 % in | 4 genes |
| 4 | 248408 | chr6 | NT_167244.1 | 3171976-3420384 | 5.12 % in | 65 repeats | 8.34 % in | 2 genes |
| 5 | 246077 | chr20 | NT_011362.10 | 31159829-31405906 | 17.01 % in | 242 repeats | 31.97 % in | 7 genes |
| 6 | 245230 | chr6 | NT_167244.1 | 4369903-4615133 | 10.62 % in | 64 repeats | 5.61 % in | 1 genes |
| 7 | 200833 | chr6 | NT_167244.1 | 3777419-3978252 | 4.25 % in | 39 repeats | 5.02 % in | 1 genes |
| 8 | 200018 | chrY | NT_011875.12 | 8474134-8674152 | 74.93 % in | 11 repeats | 0.00 % in | 0 genes |
| 9 | 191704 | chr7 | NT_007933.15 | 39730499-39922203 | 47.65 % in | 409 repeats | 0.00 % in | 0 genes |
| 10 | 181136 | chr6 | NT_167249.1 | 2137965-2319101 | 6.37 % in | 45 repeats | 0.00 % in | 0 genes |
| 11 | 180366 | chr6 | NT_167246.1 | 3056075-3236441 | 13.70 % in | 102 repeats | 0.00 % in | 0 genes |
| 12 | 175522 | chr7 | NT_023603.5 | 33558-209080 | 100.00 % in | 6 repeats | 0.00 % in | 0 genes |
| 13 | 175451 | chr16 | NT_010542.15 | 242703-418154 | 23.75 % in | 224 repeats | 0.00 % in | 0 genes |
| 14 | 174969 | chr6 | NT_167247.1 | 4419957-4594926 | 0.69 % in | 6 repeats | 0.00 % in | 0 genes |
| 15 | 171347 | chr6 | NT_167247.1 | 1561158-1732505 | 2.07 % in | 16 repeats | 0.00 % in | 0 genes |
| 16 | 167148 | chr9 | NT_008470.19 | 21685556-21852704 | 5.48 % in | 28 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 490064 | chr15 | NT_037852.6 | 1397638-1887702 | 6 | 6 | MLT1L (1) | MIRc (1) | MIRb (1) | |
| 2 | 430842 | chr6 | NT_167244.1 | 2334074-2764916 | 66 | 38 | AluSx (6) | AluJb (6) | L1ME4a (3) | |
| 3 | 271609 | chr6 | NT_167244.1 | 2003105-2274714 | 63 | 36 | AluSx (9) | MIR (5) | MIRb (3) | |
| 4 | 248408 | chr6 | NT_167244.1 | 3171976-3420384 | 65 | 28 | AluSx (10) | MIR (4) | L1MC5 (4) | |
| 5 | 246077 | chr20 | NT_011362.10 | 31159829-31405906 | 242 | 87 | MIRb (19) | MIR (16) | (TG)n (10) | |
| 6 | 245230 | chr6 | NT_167244.1 | 4369903-4615133 | 64 | 34 | HUERS-P3-int (7) | AluSx (7) | Harlequin-int (5) | |
| 7 | 200833 | chr6 | NT_167244.1 | 3777419-3978252 | 39 | 27 | L2a (6) | AT_rich (3) | MLT1H-int (2) | |
| 8 | 200018 | chrY | NT_011875.12 | 8474134-8674152 | 11 | 2 | LTR12B (10) | LTR12D (1) | ||
| 9 | 191704 | chr7 | NT_007933.15 | 39730499-39922203 | 409 | 93 | AluSx (97) | AluJo (44) | AluJb (21) | |
| 10 | 181136 | chr6 | NT_167249.1 | 2137965-2319101 | 45 | 24 | Charlie2b (6) | AluSx (5) | HAL1 (4) | |
| 11 | 180366 | chr6 | NT_167246.1 | 3056075-3236441 | 102 | 42 | AluSx (15) | AluY (12) | AluJb (7) | |
| 12 | 175522 | chr7 | NT_023603.5 | 33558-209080 | 6 | 3 | ALR/Alpha (3) | L1PA2 (2) | L1PA3 (1) | |
| 13 | 175451 | chr16 | NT_010542.15 | 242703-418154 | 224 | 68 | AluSx (30) | AluJo (16) | AluY (14) | |
| 14 | 174969 | chr6 | NT_167247.1 | 4419957-4594926 | 6 | 6 | MIR (1) | MER11A (1) | L2b (1) | |
| 15 | 171347 | chr6 | NT_167247.1 | 1561158-1732505 | 16 | 13 | MSTB (2) | MIR (2) | L1MEe (2) | |
| 16 | 167148 | chr9 | NT_008470.19 | 21685556-21852704 | 28 | 19 | L1M5 (3) | MIRb (2) | MER5B (2) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 2 | 430842 | chr6 | NT_167244.1 | 2334074-2764916 | HCG22 | HLA_complex_group_22 |
| 3 | 271609 | chr6 | NT_167244.1 | 2003105-2274714 | LOC100294090 | hypothetical_LOC100294090,_transcript_variant_1 |
| FLOT1 | flotillin-1 | |||||
| DDR1 | epithelial_discoidin_domain-containing_receptor_1_isoform_DDR1c | |||||
| MUC21 | mucin-21_precursor | |||||
| 4 | 248408 | chr6 | NT_167244.1 | 3171976-3420384 | EHMT2 | histone-lysine_N-methyltransferase,_H3_lysine-9_specific_3_isoform_b |
| TNXB | tenascin-X_isoform_1_precursor | |||||
| 5 | 246077 | chr20 | NT_011362.10 | 31159829-31405906 | CABLES2 | CDK5_and_ABL1_enzyme_substrate_2 |
| C20orf151 | hypothetical_protein_LOC140893 | |||||
| GATA5 | transcription_factor_GATA-5 | |||||
| C20orf200 | chromosome_20_open_reading_frame_200 | |||||
| MIR133A2 | microRNA:hsa-mir-133a-2 | |||||
| RPL7P3 | ||||||
| LOC100131174 | hypothetical_LOC100131174 | |||||
| 6 | 245230 | chr6 | NT_167244.1 | 4369903-4615133 | HLA-DPB2 | major_histocompatibility_complex,_class_II,_DP_beta_2_(pseudogene) |
| 7 | 200833 | chr6 | NT_167244.1 | 3777419-3978252 | HLA-DRB3 | major_histocompatibility_complex,_class_II,_DR_beta_3_precursor |