| Enzyme: | PshAI | Longest uncut segments |
| Specificity: | GACNNNNGTC | Repeats in uncut segments |
| Number of sites: | 267943 | Genes in uncut segments |
| Mean distance between sites: | 10678 base pairs | |
| Standard deviation: | 11530 base pairs | |
| Site density | 93.6 per megabase | Help |
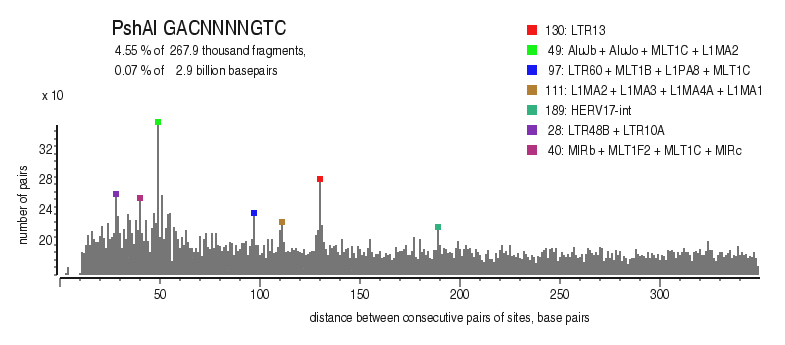
| Distribution of closely spaced sites |
|---|
 |
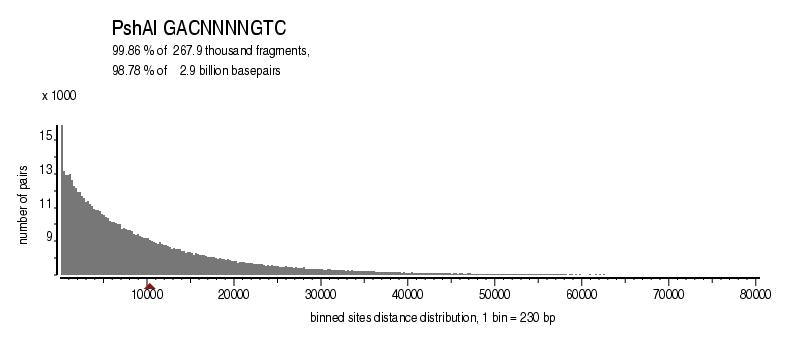
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 489822 | chr15 | NT_037852.6 | 1397448-1887270 | 0.14 % in | 6 repeats | 0.00 % in | 0 genes |
| 2 | 406116 | chr6 | NT_167244.1 | 2357747-2763863 | 0.50 % in | 9 repeats | 0.00 % in | 0 genes |
| 3 | 293721 | chr19 | NT_011109.16 | 100366-394087 | 99.76 % in | 72 repeats | 0.00 % in | 0 genes |
| 4 | 268046 | chr6 | NT_167244.1 | 2003530-2271576 | 4.51 % in | 56 repeats | 5.62 % in | 4 genes |
| 5 | 232608 | chr6 | NT_167244.1 | 4383669-4616277 | 7.87 % in | 50 repeats | 0.00 % in | 0 genes |
| 6 | 230890 | chr13 | NT_024524.14 | 31425900-31656790 | 53.53 % in | 477 repeats | 63.65 % in | 4 genes |
| 7 | 200385 | chr6 | NT_167244.1 | 3787006-3987391 | 5.04 % in | 36 repeats | 0.24 % in | 1 genes |
| 8 | 198500 | chr7 | NT_007933.15 | 26347-224847 | 99.47 % in | 55 repeats | 0.00 % in | 0 genes |
| 9 | 194721 | chr6 | NT_167249.1 | 2128124-2322845 | 10.69 % in | 74 repeats | 0.00 % in | 0 genes |
| 10 | 194230 | chr17 | NT_167251.1 | 1234852-1429082 | 37.93 % in | 346 repeats | 0.00 % in | 0 genes |
| 11 | 190322 | chr6 | NT_167247.1 | 4414480-4604802 | 3.67 % in | 32 repeats | 0.00 % in | 0 genes |
| 12 | 187673 | chr6 | NT_167244.1 | 3177148-3364821 | 2.78 % in | 32 repeats | 0.00 % in | 0 genes |
| 13 | 182365 | chrX | NT_011786.16 | 1338342-1520707 | 54.56 % in | 293 repeats | 0.00 % in | 0 genes |
| 14 | 177427 | chr12 | NT_029419.12 | 102937-280364 | 93.40 % in | 100 repeats | 0.00 % in | 0 genes |
| 15 | 176739 | chr6 | NT_167247.1 | 1562005-1738744 | 4.87 % in | 28 repeats | 0.00 % in | 0 genes |
| 16 | 171935 | chr6 | NT_167248.1 | 518797-690732 | 4.66 % in | 8 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 489822 | chr15 | NT_037852.6 | 1397448-1887270 | 6 | 6 | MLT1L (1) | MIRc (1) | MIRb (1) | |
| 2 | 406116 | chr6 | NT_167244.1 | 2357747-2763863 | 9 | 7 | L4 (2) | AluJb (2) | L1MEg (1) | |
| 3 | 293721 | chr19 | NT_011109.16 | 100366-394087 | 72 | 14 | ALR/Alpha (45) | L1PA4 (7) | L1PA3 (7) | |
| 4 | 268046 | chr6 | NT_167244.1 | 2003530-2271576 | 56 | 32 | AluSx (8) | MIR (5) | MIRb (3) | |
| 5 | 232608 | chr6 | NT_167244.1 | 4383669-4616277 | 50 | 28 | AluSx (7) | Harlequin-int (5) | HERVH-int (4) | |
| 6 | 230890 | chr13 | NT_024524.14 | 31425900-31656790 | 477 | 113 | AluSx (55) | AT_rich (29) | AluY (28) | |
| 7 | 200385 | chr6 | NT_167244.1 | 3787006-3987391 | 36 | 28 | L2a (6) | MLT1H-int (2) | L1M5 (2) | |
| 8 | 198500 | chr7 | NT_007933.15 | 26347-224847 | 55 | 12 | ALR/Alpha (30) | L1PA4 (7) | L1PA2 (5) | |
| 9 | 194721 | chr6 | NT_167249.1 | 2128124-2322845 | 74 | 36 | AluSx (8) | Charlie2b (6) | HAL1 (4) | |
| 10 | 194230 | chr17 | NT_167251.1 | 1234852-1429082 | 346 | 84 | AluSx (45) | AluJb (26) | AluJo (20) | |
| 11 | 190322 | chr6 | NT_167247.1 | 4414480-4604802 | 32 | 24 | AluSx (3) | MLT1J (2) | MIRc (2) | |
| 12 | 187673 | chr6 | NT_167244.1 | 3177148-3364821 | 32 | 18 | AluSx (6) | MIR (3) | GC_rich (3) | |
| 13 | 182365 | chrX | NT_011786.16 | 1338342-1520707 | 293 | 114 | AT_rich (20) | L2c (14) | L2a (12) | |
| 14 | 177427 | chr12 | NT_029419.12 | 102937-280364 | 100 | 42 | AluSx (18) | ALR/Alpha (10) | AluJb (6) | |
| 15 | 176739 | chr6 | NT_167247.1 | 1562005-1738744 | 28 | 18 | L1PB2 (4) | L1MEf (3) | MSTB (2) | |
| 16 | 171935 | chr6 | NT_167248.1 | 518797-690732 | 8 | 5 | AT_rich (4) | MLT1G3 (1) | L1PREC2 (1) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 4 | 268046 | chr6 | NT_167244.1 | 2003530-2271576 | LOC100294090 | hypothetical_LOC100294090,_transcript_variant_1 |
| FLOT1 | flotillin-1 | |||||
| DDR1 | epithelial_discoidin_domain-containing_receptor_1_isoform_DDR1c | |||||
| MUC21 | mucin-21_precursor | |||||
| 6 | 230890 | chr13 | NT_024524.14 | 31425900-31656790 | CTAGE10P | CTAGE_family,_member_10,_pseudogene |
| RNY4P9 | ||||||
| C13orf1 | chronic_lymphocytic_leukemia_deletion_region_gene_6_protein_isoform_2 | |||||
| RPL18P10 | deleted_in_lymphocytic_leukemia_1_(non-protein_coding) | |||||
| 7 | 200385 | chr6 | NT_167244.1 | 3787006-3987391 | HLA-DRB3 | major_histocompatibility_complex,_class_II,_DR_beta_3_precursor |