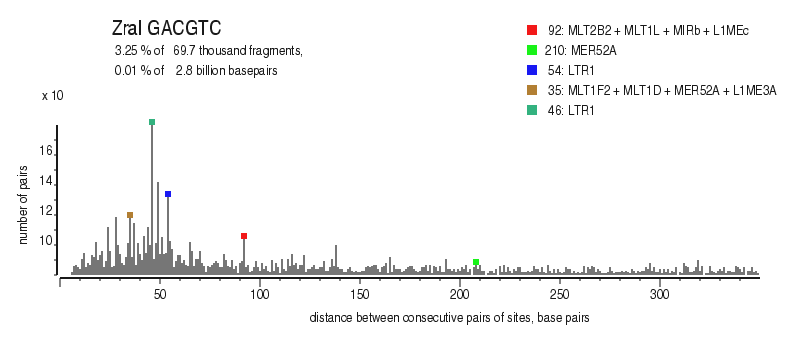
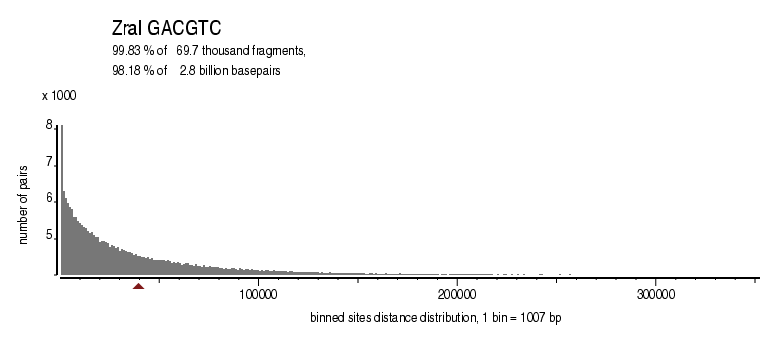
| Enzyme: | ZraI | Longest uncut segments |
| Specificity: | GACGTC | Repeats in uncut segments |
| Number of sites: | 69719 | Genes in uncut segments |
| Mean distance between sites: | 41041 base pairs | |
| Standard deviation: | 50398 base pairs | |
| Site density | 24.4 per megabase | Help |
| Distribution of closely spaced sites |
|---|
 |
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 1016518 | chr12 | NT_029419.12 | 36144301-37160819 | 56.85 % in | 1655 repeats | 16.29 % in | 4 genes |
| 2 | 824351 | chrX | NT_011651.17 | 787845-1612196 | 74.16 % in | 1042 repeats | 10.38 % in | 7 genes |
| 3 | 696592 | chr6 | NT_025741.15 | 6808601-7505193 | 56.68 % in | 1088 repeats | 0.00 % in | 0 genes |
| 4 | 609742 | chr5 | NT_023133.13 | 10123324-10733066 | 46.25 % in | 1012 repeats | 0.29 % in | 3 genes |
| 5 | 609400 | chr4 | NT_006238.11 | 5673403-6282803 | 50.32 % in | 977 repeats | 37.54 % in | 2 genes |
| 6 | 589458 | chr1 | NT_004487.19 | 25777141-26366599 | 56.77 % in | 1017 repeats | 100.00 % in | 1 genes |
| 7 | 582842 | chr3 | NT_005612.16 | 72425828-73008670 | 55.41 % in | 900 repeats | 0.09 % in | 1 genes |
| 8 | 571106 | chr3 | NT_005612.16 | 71359277-71930383 | 53.07 % in | 968 repeats | 1.74 % in | 1 genes |
| 9 | 566260 | chr15 | NT_037852.6 | 1339256-1905516 | 7.53 % in | 164 repeats | 0.00 % in | 0 genes |
| 10 | 561568 | chr6 | NT_025741.15 | 44836994-45398562 | 57.37 % in | 929 repeats | 0.00 % in | 0 genes |
| 11 | 545960 | chr2 | NT_005403.17 | 37121352-37667312 | 56.95 % in | 906 repeats | 0.00 % in | 0 genes |
| 12 | 542922 | chr10 | NT_030059.13 | 60464560-61007482 | 52.05 % in | 870 repeats | 0.00 % in | 0 genes |
| 13 | 540922 | chr12 | NT_029419.12 | 62146475-62687397 | 62.31 % in | 1087 repeats | 0.00 % in | 0 genes |
| 14 | 531120 | chr6 | NT_007299.13 | 9995305-10526425 | 47.68 % in | 870 repeats | 0.00 % in | 0 genes |
| 15 | 529994 | chr2 | NT_022135.16 | 36208774-36738768 | 45.63 % in | 779 repeats | 0.00 % in | 0 genes |
| 16 | 527971 | chr20 | NT_011362.10 | 10274174-10802145 | 55.18 % in | 1071 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 1016518 | chr12 | NT_029419.12 | 36144301-37160819 | 1655 | 359 | AT_rich (175) | L2a (53) | (TA)n (47) | |
| 2 | 824351 | chrX | NT_011651.17 | 787845-1612196 | 1042 | 218 | L2c (52) | MIRb (50) | AT_rich (43) | |
| 3 | 696592 | chr6 | NT_025741.15 | 6808601-7505193 | 1088 | 272 | AT_rich (165) | (TA)n (34) | MIR (23) | |
| 4 | 609742 | chr5 | NT_023133.13 | 10123324-10733066 | 1012 | 262 | AT_rich (73) | AluSx (53) | MIRb (44) | |
| 5 | 609400 | chr4 | NT_006238.11 | 5673403-6282803 | 977 | 247 | AT_rich (114) | MIR (59) | L2a (47) | |
| 6 | 589458 | chr1 | NT_004487.19 | 25777141-26366599 | 1017 | 218 | AT_rich (77) | AluSx (77) | MIRb (38) | |
| 7 | 582842 | chr3 | NT_005612.16 | 72425828-73008670 | 900 | 244 | AT_rich (114) | AluSx (25) | MIR (22) | |
| 8 | 571106 | chr3 | NT_005612.16 | 71359277-71930383 | 968 | 256 | AT_rich (104) | L2a (33) | MIRb (31) | |
| 9 | 566260 | chr15 | NT_037852.6 | 1339256-1905516 | 164 | 72 | L1MDa (10) | (CAGGG)n (8) | AT_rich (7) | |
| 10 | 561568 | chr6 | NT_025741.15 | 44836994-45398562 | 929 | 238 | AT_rich (89) | AluSx (26) | MIR (23) | |
| 11 | 545960 | chr2 | NT_005403.17 | 37121352-37667312 | 906 | 247 | AT_rich (71) | MIRb (27) | AluSx (27) | |
| 12 | 542922 | chr10 | NT_030059.13 | 60464560-61007482 | 870 | 243 | MIRb (63) | AT_rich (50) | MIR (33) | |
| 13 | 540922 | chr12 | NT_029419.12 | 62146475-62687397 | 1087 | 241 | AluSx (80) | AT_rich (66) | L2c (48) | |
| 14 | 531120 | chr6 | NT_007299.13 | 9995305-10526425 | 870 | 243 | AT_rich (67) | MIR (36) | L2a (33) | |
| 15 | 529994 | chr2 | NT_022135.16 | 36208774-36738768 | 779 | 215 | AT_rich (82) | MIRb (30) | MIR (28) | |
| 16 | 527971 | chr20 | NT_011362.10 | 10274174-10802145 | 1071 | 254 | MIRb (68) | AluSx (51) | MIR (47) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 1016518 | chr12 | NT_029419.12 | 36144301-37160819 | RPL31P48 | |
| LOC100128674 | hypothetical_protein_LOC100507377 | |||||
| VENTXP3 | ||||||
| ATXN7L3B | ataxin_7-like_3B | |||||
| 2 | 824351 | chrX | NT_011651.17 | 787845-1612196 | CYSLTR1 | cysteinyl_leukotriene_receptor_1 |
| ZCCHC5 | zinc_finger_CCHC_domain-containing_protein_5 | |||||
| LOC100421601 | ||||||
| LPAR4 | lysophosphatidic_acid_receptor_4 | |||||
| RPL7P54 | ||||||
| MIR4328 | microRNA_4328 | |||||
| P2RY10 | putative_P2Y_purinoceptor_10 | |||||
| 4 | 609742 | chr5 | NT_023133.13 | 10123324-10733066 | RPL21P59 | |
| RPL7P20 | ||||||
| RPLP0P9 | ||||||
| 5 | 609400 | chr4 | NT_006238.11 | 5673403-6282803 | GABRG1 | gamma-aminobutyric_acid_receptor_subunit_gamma-1_precursor |
| GABRA2 | gamma-aminobutyric_acid_receptor_subunit_alpha-2_precursor | |||||
| 6 | 589458 | chr1 | NT_004487.19 | 25777141-26366599 | RPS26P12 | rab_GTPase-activating_protein_1-like_isoform_B |
| 7 | 582842 | chr3 | NT_005612.16 | 72425828-73008670 | LOC100287630 | |
| 8 | 571106 | chr3 | NT_005612.16 | 71359277-71930383 | SLITRK3 | SLIT_and_NTRK-like_protein_3_precursor |