| Enzyme: | AflII | Longest uncut segments |
| Specificity: | CTTAAG | Repeats in uncut segments |
| Number of sites: | 640622 | Genes in uncut segments |
| Mean distance between sites: | 4466 base pairs | |
| Standard deviation: | 4755 base pairs | |
| Site density | 223.9 per megabase | Help |
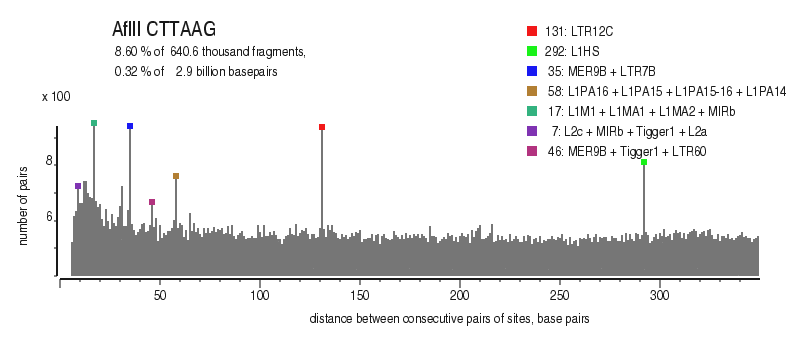
| Distribution of closely spaced sites |
|---|
 |
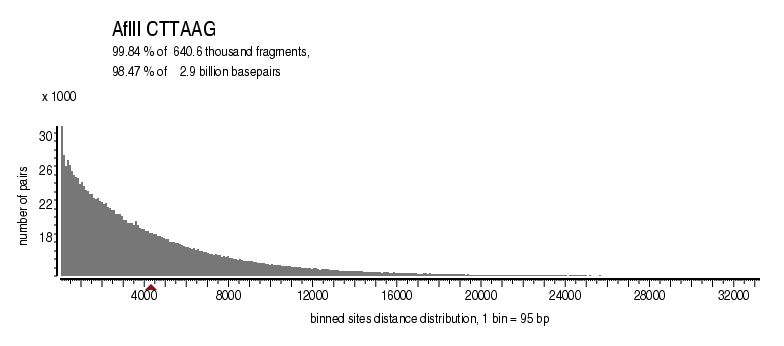
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 492230 | chr15 | NT_037852.6 | 1396227-1888457 | 0.44 % in | 12 repeats | 0.00 % in | 0 genes |
| 2 | 413086 | chr6 | NT_167244.1 | 2359198-2772284 | 1.16 % in | 20 repeats | 0.00 % in | 0 genes |
| 3 | 267200 | chr6 | NT_167244.1 | 1991513-2258713 | 3.78 % in | 49 repeats | 6.82 % in | 4 genes |
| 4 | 231561 | chr6 | NT_167244.1 | 4374636-4606197 | 7.25 % in | 36 repeats | 3.89 % in | 1 genes |
| 5 | 190503 | chr6 | NT_167244.1 | 3178729-3369232 | 2.74 % in | 24 repeats | 7.33 % in | 2 genes |
| 6 | 188915 | chr6 | NT_167244.1 | 3787262-3976177 | 2.57 % in | 22 repeats | 0.12 % in | 1 genes |
| 7 | 183481 | chr6 | NT_167247.1 | 4416310-4599791 | 2.41 % in | 22 repeats | 99.56 % in | 1 genes |
| 8 | 176906 | chr6 | NT_167247.1 | 1550352-1727258 | 4.12 % in | 37 repeats | 7.12 % in | 1 genes |
| 9 | 176564 | chr6 | NT_167249.1 | 2129007-2305571 | 4.44 % in | 32 repeats | 0.00 % in | 0 genes |
| 10 | 163616 | chr6 | NT_167244.1 | 2890043-3053659 | 5.21 % in | 44 repeats | 0.00 % in | 0 genes |
| 11 | 162492 | chr6 | NT_167248.1 | 519493-681985 | 1.96 % in | 2 repeats | 0.00 % in | 0 genes |
| 12 | 161945 | chr4 | NT_006316.16 | 390823-552768 | 3.35 % in | 56 repeats | 0.00 % in | 0 genes |
| 13 | 159617 | chr9 | NT_008470.19 | 21690884-21850501 | 2.20 % in | 17 repeats | 0.00 % in | 0 genes |
| 14 | 143839 | chr1 | NT_167185.1 | 1489610-1633449 | 8.07 % in | 49 repeats | 0.00 % in | 0 genes |
| 15 | 135071 | chr1 | NT_167185.1 | 30282-165353 | 11.61 % in | 55 repeats | 0.00 % in | 0 genes |
| 16 | 128323 | chr6 | NT_167245.1 | 2598077-2726400 | 6.82 % in | 20 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 492230 | chr15 | NT_037852.6 | 1396227-1888457 | 12 | 11 | L2a (2) | (TA)n (1) | MLT1L (1) | |
| 2 | 413086 | chr6 | NT_167244.1 | 2359198-2772284 | 20 | 14 | AluY (3) | LTR84b (2) | L4 (2) | |
| 3 | 267200 | chr6 | NT_167244.1 | 1991513-2258713 | 49 | 29 | AluSx (7) | L2c (4) | MIRb (3) | |
| 4 | 231561 | chr6 | NT_167244.1 | 4374636-4606197 | 36 | 22 | HUERS-P3-int (5) | HERVH-int (4) | AluSx (4) | |
| 5 | 190503 | chr6 | NT_167244.1 | 3178729-3369232 | 24 | 13 | AluSx (4) | MIR (3) | GC_rich (3) | |
| 6 | 188915 | chr6 | NT_167244.1 | 3787262-3976177 | 22 | 18 | L2a (3) | MLT1H-int (2) | AT_rich (2) | |
| 7 | 183481 | chr6 | NT_167247.1 | 4416310-4599791 | 22 | 18 | AluSx (3) | MLT1J (2) | L1MC5 (2) | |
| 8 | 176906 | chr6 | NT_167247.1 | 1550352-1727258 | 37 | 25 | MIRc (3) | MIR3 (3) | L2c (3) | |
| 9 | 176564 | chr6 | NT_167249.1 | 2129007-2305571 | 32 | 17 | AluSx (5) | MamGypLTR1b (3) | L1MB8 (3) | |
| 10 | 163616 | chr6 | NT_167244.1 | 2890043-3053659 | 44 | 23 | L1MC5 (6) | AluY (5) | AluSc (4) | |
| 11 | 162492 | chr6 | NT_167248.1 | 519493-681985 | 2 | 2 | L1PREC2 (1) | HERVH-int (1) | ||
| 12 | 161945 | chr4 | NT_006316.16 | 390823-552768 | 56 | 4 | (CA)n (47) | L1M4 (7) | MER5B (1) | |
| 13 | 159617 | chr9 | NT_008470.19 | 21690884-21850501 | 17 | 13 | MIRb (2) | LTR67B (2) | L2 (2) | |
| 14 | 143839 | chr1 | NT_167185.1 | 1489610-1633449 | 49 | 21 | GA-rich (5) | MIRc (4) | (GA)n (4) | |
| 15 | 135071 | chr1 | NT_167185.1 | 30282-165353 | 55 | 35 | AluY (4) | (TG)n (3) | L1PB1 (3) | |
| 16 | 128323 | chr6 | NT_167245.1 | 2598077-2726400 | 20 | 17 | MLT1N2 (2) | MER21C (2) | L2 (2) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 3 | 267200 | chr6 | NT_167244.1 | 1991513-2258713 | MDC1 | mediator_of_DNA_damage_checkpoint_protein_1 |
| LOC100294090 | hypothetical_LOC100294090,_transcript_variant_1 | |||||
| FLOT1 | flotillin-1 | |||||
| DDR1 | epithelial_discoidin_domain-containing_receptor_1_isoform_DDR1c | |||||
| 4 | 231561 | chr6 | NT_167244.1 | 4374636-4606197 | HLA-DPB2 | major_histocompatibility_complex,_class_II,_DP_beta_2_(pseudogene) |
| 5 | 190503 | chr6 | NT_167244.1 | 3178729-3369232 | EHMT2 | histone-lysine_N-methyltransferase,_H3_lysine-9_specific_3_isoform_b |
| TNXB | tenascin-X_isoform_1_precursor | |||||
| 6 | 188915 | chr6 | NT_167244.1 | 3787262-3976177 | HLA-DRB3 | major_histocompatibility_complex,_class_II,_DR_beta_3_precursor |
| 7 | 183481 | chr6 | NT_167247.1 | 4416310-4599791 | LOC100507722 | hypothetical_protein_LOC100507722 |
| 8 | 176906 | chr6 | NT_167247.1 | 1550352-1727258 | LOC100421582 | tripartite_motif-containing_protein_26 |