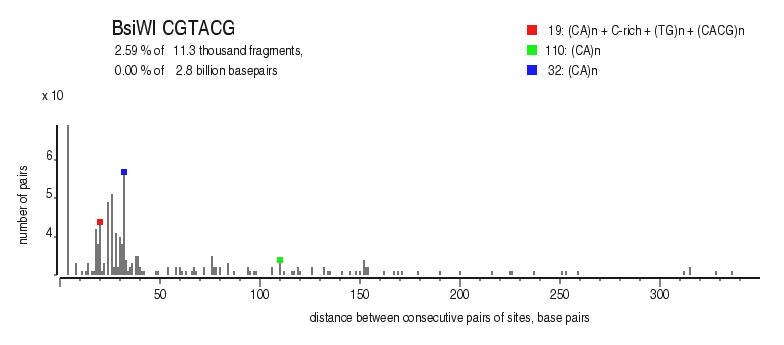
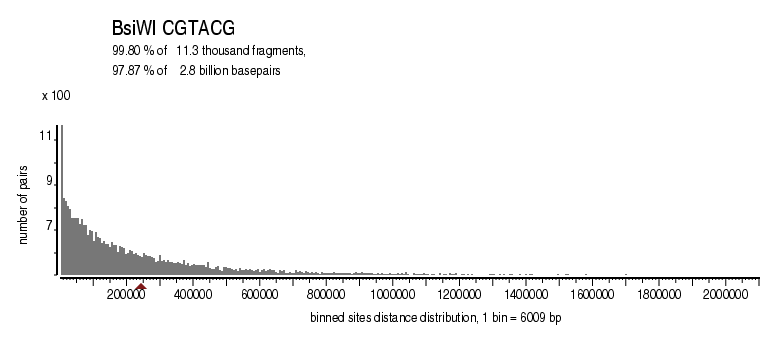
|
Help |
|
|---|
Genes in longest uncut segments
|
|---|
| Sgmnt |
Length (bp) |
Chr |
Scaffold |
Coordinates |
Gene symbol |
Gene function |
|---|
| 1 |
3704546 |
chr6 |
NT_007299.13 |
13754805-17459351 |
COL12A1 |
collagen_alpha-1(XII)_chain_short_isoform_precursor |
|
| COX7A2 |
cytochrome_c_oxidase_subunit_7A2,_mitochondrial_precursor |
|
| TMEM30A |
cell_cycle_control_protein_50A_isoform_2 |
|
| LOC100506804 |
hypothetical_LOC100506804,_transcript_variant_2 |
|
| LOC100288170 |
filamin-A-interacting_protein_1 |
|
| RPL26P20 |
|
|
| LOC100421091 |
sentrin-specific_protease_6_isoform_2 |
|
| MYO6 |
myosin-VI |
|
| IMPG1 |
interphotoreceptor_matrix_proteoglycan_1_precursor |
|
| LOC100131680 |
|
|
| HTR1B |
5-hydroxytryptamine_receptor_1B |
|
| RPS6P7 |
|
| 2 |
3160673 |
chr1 |
NT_032977.9 |
48483273-51643946 |
LOC100422537 |
PDZ_domain-containing_protein_GIPC2 |
|
| LOC100132264 |
|
|
| MGC27382 |
hypothetical_MGC27382 |
|
| PTGFR |
prostaglandin_F2-alpha_receptor_isoform_b_precursor |
|
| IFI44L |
interferon-induced_protein_44-like |
|
| IFI44 |
interferon-induced_protein_44 |
|
| RPL23P3 |
|
|
| LOC652549 |
|
|
| ELTD1 |
EGF,_latrophilin_and_seven_transmembrane_domain-containing_protein_1_precursor |
|
| LOC729779 |
|
|
| ADH5P2 |
|
|
| LOC553139 |
|
|
| HMGB1P18 |
|
|
| LOC100129325 |
|
|
| LOC100129325 |
|
|
| RPL7P10 |
|
| 3 |
3084035 |
chr6 |
NT_025741.15 |
16774519-19858554 |
LAMA4 |
laminin_subunit_alpha-4_isoform_3_precursor |
|
| LOC100128588 |
|
|
| RFPL4B |
ret_finger_protein-like_4B |
|
| RPSAP45 |
|
|
| LOC442249 |
|
|
| LOC643859 |
|
|
| PA2G4P5 |
|
|
| LOC100287612 |
hypothetical_LOC100287612 |
|
| LOC643884 |
|
|
| RPS27AP11 |
|
|
| RPL30P8 |
|
|
| MARCKS |
myristoylated_alanine-rich_C-kinase_substrate |
|
| FLJ34503 |
hypothetical_FLJ34503 |
|
| HDAC2 |
histone_deacetylase_2 |
|
| LOC100422450 |
|
|
| HS3ST5 |
heparan_sulfate_glucosamine_3-O-sulfotransferase_5 |
|
| RPSAP43 |
|
|
| LOC441167 |
hCG1820801 |
|
| LOC728614 |
|
| 4 |
2911707 |
chr11 |
NT_033899.8 |
7870462-10782169 |
LOC100506742 |
inactive_caspase-12-like_isoform_2 |
|
| LOC643733 |
caspase_4,_apoptosis-related_cysteine_peptidase_pseudogene,_transcript_variant_2 |
|
| CASP4 |
caspase-4_isoform_gamma_precursor |
|
| CASP5 |
caspase-5_isoform_f_precursor |
|
| CASP1 |
caspase-1_isoform_gamma_precursor |
|
| CARD16 |
caspase_recruitment_domain-containing_protein_16_isoform_2 |
|
| LOC440067 |
|
|
| CARD17 |
caspase_recruitment_domain-containing_protein_17 |
|
| CARD18 |
caspase_recruitment_domain-containing_protein_18 |
|
| OR2AL1P |
|
|
| HSPD1P13 |
glutamate_receptor_4_isoform_3_precursor |
|
| KIAA1826 |
hypothetical_protein_LOC84437 |
|
| KBTBD3 |
kelch_repeat_and_BTB_domain-containing_protein_3 |
|
| AASDHPPT |
L-aminoadipate-semialdehyde_dehydrogenase-phosphopantetheinyl_transferase |
|
| LOC643855 |
|
|
| LOC100422300 |
guanylate_cyclase_soluble_subunit_alpha-2 |
|
| ASS1P13 |
|
|
| LOC100271884 |
CWF19-like_protein_2 |
| 5 |
2875121 |
chr4 |
NT_016354.19 |
80635202-83510323 |
NPY2R |
neuropeptide_Y_receptor_type_2 |
|
| MAP9 |
microtubule-associated_protein_9 |
|
| LOC100420289 |
|
|
| LOC100287939 |
|
|
| TRNAQ54P |
|
|
| LOC100287591 |
|
|
| - |
tRNA-Leu |
|
| GUCY1A3 |
guanylate_cyclase_soluble_subunit_alpha-3_isoform_B |
|
| GUCY1B3 |
guanylate_cyclase_soluble_subunit_beta-1 |
|
| ACCN5 |
amiloride-sensitive_cation_channel_5 |
|
| TDO2 |
tryptophan_2,3-dioxygenase |
|
| CTSO |
cathepsin_O_preproprotein_preproprotein |
|
| FTH1P21 |
|
|
| LOC100505784 |
hypothetical_LOC100505784 |
|
| PDGFC |
platelet-derived_growth_factor_C_precursor |
|
| GLRB |
glycine_receptor_subunit_beta_isoform_B_precursor |
|
| LOC391707 |
|
|
| GRIA2 |
glutamate_receptor_2_isoform_3 |
|
| LOC340017 |
hypothetical_LOC340017 |
|
| RPL6P11 |
|
| 6 |
2747548 |
chr3 |
NT_005612.16 |
20672664-23420212 |
LOC100506673 |
hypothetical_LOC100506673 |
|
| LOC645207 |
|
|
| GAP43 |
neuromodulin_isoform_1 |
|
| LOC100506708 |
hypothetical_LOC100506708 |
|
| BZW1P2 |
|
|
| LOC285194 |
hypothetical_LOC285194 |
|
| LOC100506724 |
hypothetical_LOC100506724 |
|
| LOC728873 |
|
| 7 |
2733096 |
chr1 |
NT_167186.1 |
14101513-16834609 |
LOC100129664 |
|
|
| LOC100421052 |
|
|
| LOC100419489 |
serine/threonine-protein_kinase_MARK1 |
|
| C1orf115 |
hypothetical_protein_LOC79762 |
|
| MOSC2 |
MOSC_domain-containing_protein_2,_mitochondrial_precursor |
|
| MOSC1 |
MOSC_domain-containing_protein_1,_mitochondrial_precursor |
|
| HLX |
H2.0-like_homeobox_protein |
|
| LOC100132626 |
|
|
| LOC400804 |
hypothetical_LOC400804 |
|
| LOC100132179 |
|
|
| DUSP10 |
dual_specificity_protein_phosphatase_10_isoform_b |
|
| LOC100422330 |
|
|
| - |
tRNA-Thr |
|
| CICP13 |
|
|
| LOC653056 |
|
|
| LOC653056 |
hypothetical_LOC728417 |
|
| HHIPL2 |
HHIP-like_protein_2_precursor |
|
| TAF1A |
TATA_box-binding_protein-associated_factor_RNA_polymerase_I_subunit_A_isoform_2 |
|
| LOC100506161 |
hypothetical_LOC100506161 |
|
| LOC100291899 |
melanoma_inhibitory_activity_protein_3_precursor |
|
| AIDA |
axin_interactor,_dorsalization-associated_protein |
|
| C1orf58 |
BRO1_domain-containing_protein_BROX |
|
| FAM177B |
hypothetical_protein_LOC400823 |
|
| NDUFB1P2 |
protein_dispatched_homolog_1 |
|
| TLR5 |
toll-like_receptor_5_precursor |
| 8 |
2680511 |
chr3 |
NT_005612.16 |
2774969-5455480 |
LOC391556 |
|
|
| MTRNR2L12 |
|
|
| RPL18AP8 |
|
|
| LOC100131442 |
|
|
| LOC100129736 |
|
|
| EPHA6 |
ephrin_type-A_receptor_6_isoform_b |
|
| ARL6 |
ADP-ribosylation_factor-like_protein_6 |
|
| LOC100506362 |
hypothetical_LOC100506362 |
|
| CRYBG3 |
hypothetical_protein_LOC131544 |
|
| MINA |
MYC-induced_nuclear_antigen_isoform_b |
|
| GABRR3 |
gamma-aminobutyric_acid_receptor_subunit_rho-3_precursor |
|
| OR5BM1P |
|
|
| OR5AC1 |
|
|
| OR5AC2 |
olfactory_receptor_5AC2 |
|
| OR5AC4P |
|
|
| POU5F1P7 |
|
|
| OR5H1 |
olfactory_receptor_5H1 |
|
| OR5H14 |
olfactory_receptor_5H14 |
|
| OR5H15 |
olfactory_receptor_5H15 |
|
| OR5H5P |
|
|
| OR5H3P |
|
|
| OR5H4P |
|
|
| OR5H7P |
|
|
| OR5H6 |
olfactory_receptor_5H6 |
|
| OR5H2 |
olfactory_receptor_5H2 |
|
| OR5H8P |
|
|
| OR5K4 |
olfactory_receptor_5K4 |
|
| OR5K3 |
olfactory_receptor_5K3 |
|
| LOC100130484 |
|
|
| OR5K1 |
olfactory_receptor_5K1 |
|
| OR5K2 |
olfactory_receptor_5K2 |
|
| CLDND1 |
claudin_domain-containing_protein_1_isoform_a |
|
| LOC100130963 |
|
|
| GPR15 |
G-protein_coupled_receptor_15 |
|
| CPOX |
coproporphyrinogen-III_oxidase,_mitochondrial_precursor |
|
| RPL19P7 |
|
|
| LOC339843 |
|
|
| LOC100419185 |
type_2_lactosamine_alpha-2,3-sialyltransferase |
|
| DCBLD2 |
discoidin,_CUB_and_LCCL_domain-containing_protein_2_precursor |
|
| LOC100506377 |
hypothetical_LOC100506377 |
|
| LOC100418920 |
|
|