| Enzyme: | RpaB5I | Longest uncut segments |
| Specificity: | CGRGGAC | Repeats in uncut segments |
| Number of sites: | 121052 | Genes in uncut segments |
| Mean distance between sites: | 23637 base pairs | |
| Standard deviation: | 37962 base pairs | |
| Site density | 42.3 per megabase | Help |
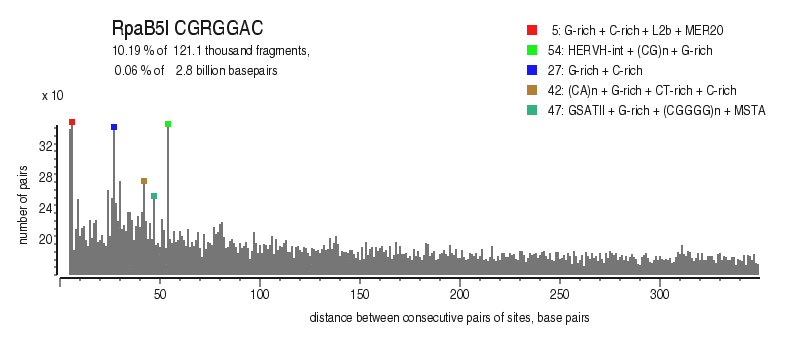
| Distribution of closely spaced sites |
|---|
 |
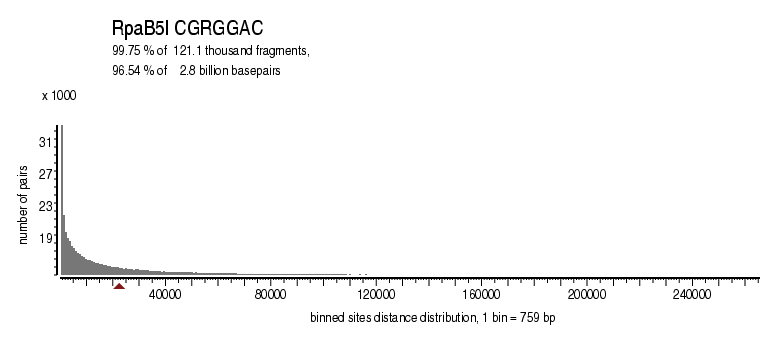
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 590132 | chr5 | NT_006713.15 | 39115362-39705494 | 44.63 % in | 856 repeats | 0.00 % in | 0 genes |
| 2 | 579110 | chr15 | NT_037852.6 | 1395277-1974387 | 6.70 % in | 167 repeats | 3.14 % in | 2 genes |
| 3 | 557220 | chr7 | NT_007819.17 | 12716484-13273704 | 44.21 % in | 924 repeats | 0.73 % in | 1 genes |
| 4 | 551627 | chr11 | NT_167190.1 | 33472812-34024439 | 50.46 % in | 929 repeats | 87.18 % in | 1 genes |
| 5 | 545646 | chr6 | NT_025741.15 | 6058546-6604192 | 36.16 % in | 851 repeats | 100.00 % in | 1 genes |
| 6 | 533152 | chr3 | NT_022517.18 | 19410690-19943842 | 61.12 % in | 858 repeats | 33.66 % in | 4 genes |
| 7 | 532463 | chr6 | NT_025741.15 | 7490452-8022915 | 55.78 % in | 847 repeats | 0.00 % in | 0 genes |
| 8 | 515543 | chr1 | NT_004487.19 | 47927468-48443011 | 51.24 % in | 747 repeats | 68.76 % in | 8 genes |
| 9 | 511557 | chr7 | NT_007933.15 | 55638011-56149568 | 49.89 % in | 857 repeats | 0.00 % in | 0 genes |
| 10 | 507158 | chr4 | NT_016354.19 | 30957478-31464636 | 47.58 % in | 827 repeats | 0.00 % in | 0 genes |
| 11 | 505900 | chr11 | NT_167190.1 | 30139510-30645410 | 67.26 % in | 839 repeats | 0.00 % in | 0 genes |
| 12 | 498221 | chr4 | NT_016354.19 | 42699036-43197257 | 55.86 % in | 742 repeats | 0.00 % in | 0 genes |
| 13 | 493379 | chr2 | NT_005403.17 | 65084016-65577395 | 46.57 % in | 745 repeats | 0.00 % in | 0 genes |
| 14 | 482572 | chr13 | NT_024524.14 | 43999928-44482500 | 49.09 % in | 752 repeats | 0.00 % in | 0 genes |
| 15 | 468453 | chr6 | NT_007592.15 | 55077856-55546309 | 48.28 % in | 682 repeats | 0.00 % in | 0 genes |
| 16 | 463049 | chr4 | NT_022778.16 | 6798868-7261917 | 56.81 % in | 713 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 590132 | chr5 | NT_006713.15 | 39115362-39705494 | 856 | 223 | AT_rich (90) | MIR (42) | L2c (42) | |
| 2 | 579110 | chr15 | NT_037852.6 | 1395277-1974387 | 167 | 72 | AT_rich (14) | AluSx (12) | AluJb (9) | |
| 3 | 557220 | chr7 | NT_007819.17 | 12716484-13273704 | 924 | 237 | AT_rich (91) | AluY (45) | AluSx (39) | |
| 4 | 551627 | chr11 | NT_167190.1 | 33472812-34024439 | 929 | 219 | MIRb (82) | L2c (63) | AT_rich (61) | |
| 5 | 545646 | chr6 | NT_025741.15 | 6058546-6604192 | 851 | 194 | AT_rich (117) | L2a (43) | MIR (41) | |
| 6 | 533152 | chr3 | NT_022517.18 | 19410690-19943842 | 858 | 229 | AT_rich (51) | MIRb (44) | AluSx (35) | |
| 7 | 532463 | chr6 | NT_025741.15 | 7490452-8022915 | 847 | 251 | AT_rich (109) | L2a (21) | L1MEc (18) | |
| 8 | 515543 | chr1 | NT_004487.19 | 47927468-48443011 | 747 | 192 | AT_rich (90) | AluSx (31) | AluY (20) | |
| 9 | 511557 | chr7 | NT_007933.15 | 55638011-56149568 | 857 | 258 | AT_rich (84) | L2c (32) | L2a (28) | |
| 10 | 507158 | chr4 | NT_016354.19 | 30957478-31464636 | 827 | 202 | AT_rich (64) | L2a (44) | MIRb (35) | |
| 11 | 505900 | chr11 | NT_167190.1 | 30139510-30645410 | 839 | 222 | MIRb (61) | AT_rich (49) | L2a (48) | |
| 12 | 498221 | chr4 | NT_016354.19 | 42699036-43197257 | 742 | 237 | AT_rich (91) | L2a (23) | L2 (20) | |
| 13 | 493379 | chr2 | NT_005403.17 | 65084016-65577395 | 745 | 212 | AT_rich (75) | L2a (41) | MIR (33) | |
| 14 | 482572 | chr13 | NT_024524.14 | 43999928-44482500 | 752 | 235 | AT_rich (127) | MIR (26) | L2a (25) | |
| 15 | 468453 | chr6 | NT_007592.15 | 55077856-55546309 | 682 | 196 | AT_rich (82) | MIRb (28) | L2c (23) | |
| 16 | 463049 | chr4 | NT_022778.16 | 6798868-7261917 | 713 | 218 | AT_rich (67) | MIR (20) | (TA)n (18) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 2 | 579110 | chr15 | NT_037852.6 | 1395277-1974387 | LOC100418897 | |
| LOC646214 | p21_protein_(Cdc42/Rac)-activated_kinase_2_pseudogene | |||||
| 3 | 557220 | chr7 | NT_007819.17 | 12716484-13273704 | ARL4A | ADP-ribosylation_factor-like_protein_4A |
| 4 | 551627 | chr11 | NT_167190.1 | 33472812-34024439 | GRM5 | metabotropic_glutamate_receptor_5_isoform_b_precursor |
| 5 | 545646 | chr6 | NT_025741.15 | 6058546-6604192 | LOC100418924 | glutamate_receptor,_ionotropic_kainate_2_isoform_2_precursor |
| 6 | 533152 | chr3 | NT_022517.18 | 19410690-19943842 | KCNH8 | potassium_voltage-gated_channel_subfamily_H_member_8 |
| EFHB | EF-hand_domain-containing_family_member_B | |||||
| LOC402125 | ||||||
| RAB5A | ras-related_protein_Rab-5A | |||||
| 8 | 515543 | chr1 | NT_004487.19 | 47927468-48443011 | KCNT2 | potassium_channel_subfamily_T_member_2 |
| CFH | complement_factor_H_isoform_b_precursor | |||||
| CFHR3 | complement_factor_H-related_protein_3_isoform_2_precursor | |||||
| CFHR1 | complement_factor_H-related_protein_1_precursor | |||||
| LOC100289145 | ||||||
| CFHR4 | complement_factor_H-related_protein_4_precursor | |||||
| CFHR2 | complement_factor_H-related_protein_2_precursor | |||||
| CFHR5 | complement_factor_H-related_protein_5_precursor | |||||