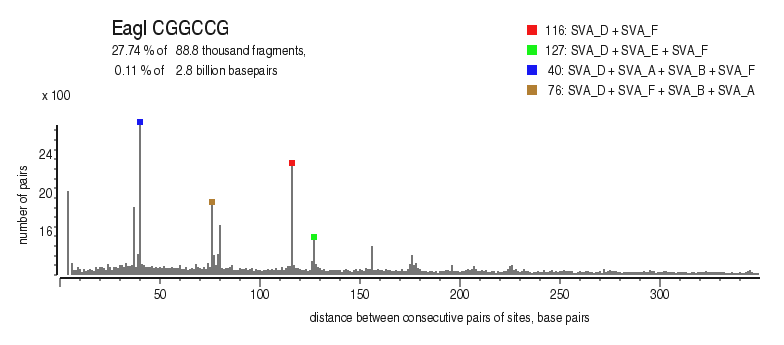
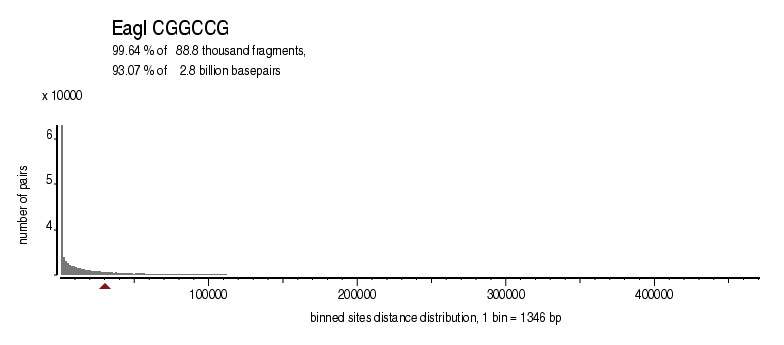
| Enzyme: | EagI | Longest uncut segments |
| Specificity: | CGGCCG | Repeats in uncut segments |
| Number of sites: | 88797 | Genes in uncut segments |
| Mean distance between sites: | 32223 base pairs | |
| Standard deviation: | 67311 base pairs | |
| Site density | 31.0 per megabase | Help |
| Distribution of closely spaced sites |
|---|
 |
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 1361329 | chrX | NT_011651.17 | 3902105-5263434 | 79.74 % in | 2003 repeats | 0.47 % in | 3 genes |
| 2 | 1229124 | chr6 | NT_007299.13 | 403999-1633123 | 50.58 % in | 1829 repeats | 49.40 % in | 3 genes |
| 3 | 1222923 | chrX | NT_011651.17 | 10074994-11297917 | 71.32 % in | 1913 repeats | 12.39 % in | 5 genes |
| 4 | 1138599 | chr10 | NT_030059.13 | 8719650-9858249 | 54.31 % in | 1794 repeats | 0.34 % in | 1 genes |
| 5 | 1113826 | chr7 | NT_007933.15 | 56571809-57685635 | 59.13 % in | 1704 repeats | 0.08 % in | 1 genes |
| 6 | 1055571 | chr2 | NT_022135.16 | 35856414-36911985 | 43.31 % in | 1547 repeats | 0.27 % in | 3 genes |
| 7 | 1036041 | chr13 | NT_024524.14 | 49536546-50572587 | 50.79 % in | 1661 repeats | 0.21 % in | 3 genes |
| 8 | 977746 | chr5 | NT_029289.11 | 5048547-6026293 | 61.19 % in | 1686 repeats | 1.72 % in | 3 genes |
| 9 | 974637 | chr4 | NT_016354.19 | 58702452-59677089 | 54.78 % in | 1417 repeats | 0.00 % in | 0 genes |
| 10 | 953105 | chr11 | NT_167190.1 | 32112530-33065635 | 55.09 % in | 1562 repeats | 0.00 % in | 0 genes |
| 11 | 935146 | chr4 | NT_016354.19 | 85778403-86713549 | 53.52 % in | 1462 repeats | 0.00 % in | 0 genes |
| 12 | 924967 | chr2 | NT_022184.15 | 38141889-39066856 | 40.89 % in | 1396 repeats | 0.00 % in | 0 genes |
| 13 | 922233 | chr10 | NT_030059.13 | 59729186-60651419 | 54.71 % in | 1553 repeats | 0.00 % in | 0 genes |
| 14 | 914477 | chr14 | NT_026437.12 | 25010783-25925260 | 55.70 % in | 1447 repeats | 0.00 % in | 0 genes |
| 15 | 913207 | chr5 | NT_034772.6 | 281011-1194218 | 41.65 % in | 1239 repeats | 0.00 % in | 0 genes |
| 16 | 908117 | chrY | NT_011875.12 | 7905471-8813588 | 79.17 % in | 850 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 1361329 | chrX | NT_011651.17 | 3902105-5263434 | 2003 | 354 | AT_rich (115) | (TA)n (49) | MIR (49) | |
| 2 | 1229124 | chr6 | NT_007299.13 | 403999-1633123 | 1829 | 341 | AT_rich (220) | MIR (75) | L2a (65) | |
| 3 | 1222923 | chrX | NT_011651.17 | 10074994-11297917 | 1913 | 370 | AT_rich (148) | AluSx (57) | L2a (40) | |
| 4 | 1138599 | chr10 | NT_030059.13 | 8719650-9858249 | 1794 | 378 | AT_rich (193) | MIR (65) | MIRb (60) | |
| 5 | 1113826 | chr7 | NT_007933.15 | 56571809-57685635 | 1704 | 351 | AT_rich (191) | (TA)n (43) | L1M5 (38) | |
| 6 | 1055571 | chr2 | NT_022135.16 | 35856414-36911985 | 1547 | 329 | AT_rich (173) | MIR (57) | MIRb (56) | |
| 7 | 1036041 | chr13 | NT_024524.14 | 49536546-50572587 | 1661 | 357 | AT_rich (216) | L2a (52) | MIR (42) | |
| 8 | 977746 | chr5 | NT_029289.11 | 5048547-6026293 | 1686 | 301 | MIRb (113) | L2a (108) | AT_rich (99) | |
| 9 | 974637 | chr4 | NT_016354.19 | 58702452-59677089 | 1417 | 332 | AT_rich (204) | L2a (47) | (TA)n (32) | |
| 10 | 953105 | chr11 | NT_167190.1 | 32112530-33065635 | 1562 | 318 | MIRb (110) | AT_rich (88) | MIR (76) | |
| 11 | 935146 | chr4 | NT_016354.19 | 85778403-86713549 | 1462 | 324 | AT_rich (198) | (TA)n (42) | L2a (38) | |
| 12 | 924967 | chr2 | NT_022184.15 | 38141889-39066856 | 1396 | 296 | AT_rich (101) | MIRb (81) | MIR (71) | |
| 13 | 922233 | chr10 | NT_030059.13 | 59729186-60651419 | 1553 | 303 | AT_rich (87) | MIRb (78) | MIR (70) | |
| 14 | 914477 | chr14 | NT_026437.12 | 25010783-25925260 | 1447 | 344 | AT_rich (171) | L2a (46) | AluSx (40) | |
| 15 | 913207 | chr5 | NT_034772.6 | 281011-1194218 | 1239 | 281 | AT_rich (147) | MIRb (60) | L2c (55) | |
| 16 | 908117 | chrY | NT_011875.12 | 7905471-8813588 | 850 | 225 | AT_rich (59) | AluY (36) | L1M1 (22) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 1361329 | chrX | NT_011651.17 | 3902105-5263434 | LOC100422471 | |
| LOC100129843 | ||||||
| RPL22P22 | ||||||
| 2 | 1229124 | chr6 | NT_007299.13 | 403999-1633123 | MTRNR2L9 | MTRNR2-like_9 |
| KHDRBS2 | KH_domain-containing,_RNA-binding,_signal_transduction-associated_protein_2 | |||||
| LOC100132056 | ||||||
| 3 | 1222923 | chrX | NT_011651.17 | 10074994-11297917 | KLHL4 | kelch-like_protein_4_isoform_1 |
| RPSAP15 | ||||||
| MRPS22P1 | ||||||
| LOC100129133 | ||||||
| CAPZA1P | ||||||
| 4 | 1138599 | chr10 | NT_030059.13 | 8719650-9858249 | ZWINT | ZW10_interactor_isoform_b |
| 5 | 1113826 | chr7 | NT_007933.15 | 56571809-57685635 | LOC648442 | |
| 6 | 1055571 | chr2 | NT_022135.16 | 35856414-36911985 | LOC727713 | |
| RNU7-2P | ||||||
| RPL17P12 | ||||||
| 7 | 1036041 | chr13 | NT_024524.14 | 49536546-50572587 | RPL37P21 | |
| RPL12P34 | ||||||
| LOC730239 | ||||||
| 8 | 977746 | chr5 | NT_029289.11 | 5048547-6026293 | LOC100128121 | |
| LOC100132712 | ||||||
| ASS1P10 | ||||||