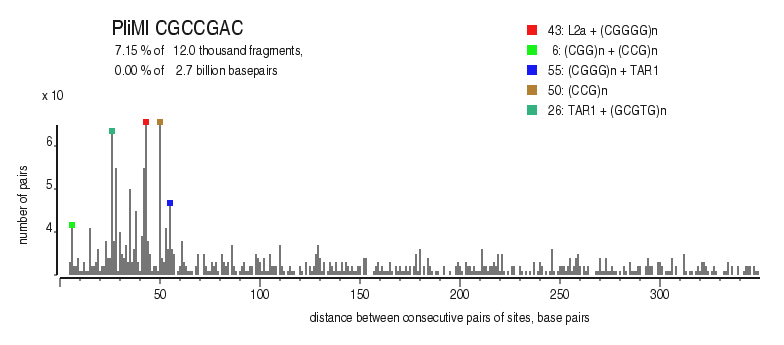
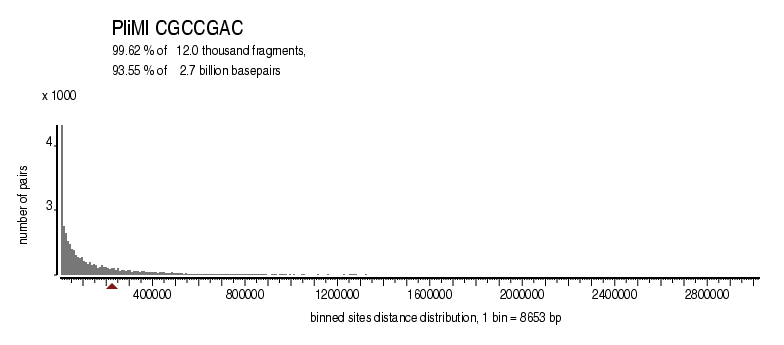
|
Help |
|
|---|
Genes in longest uncut segments
|
|---|
| Sgmnt |
Length (bp) |
Chr |
Scaffold |
Coordinates |
Gene symbol |
Gene function |
|---|
| 1 |
7052207 |
chr12 |
NT_029419.12 |
47629881-54682088 |
LRRIQ1 |
leucine-rich_repeat_and_IQ_domain-containing_protein_1_isoform_2 |
|
| ALX1 |
ALX_homeobox_protein_1 |
|
| LOC441643 |
|
|
| LOC100129589 |
|
|
| RASSF9 |
ras_association_domain-containing_protein_9 |
|
| NTS |
neurotensin/neuromedin_N_preproprotein_preproprotein |
|
| MGAT4C |
alpha-1,3-mannosyl-glycoprotein_4-beta-N-acetylglucosaminyltransferase_C |
|
| RPL23AP68 |
|
|
| LOC100507559 |
hypothetical_LOC100507559 |
|
| CYCSP30 |
|
|
| LOC100420357 |
makorin_ring_finger_protein_9,_pseudogene |
|
| RPS4XP15 |
|
|
| C12orf50 |
hypothetical_protein_LOC160419 |
|
| C12orf29 |
hypothetical_protein_LOC91298 |
|
| LOC100420011 |
centrosomal_protein_of_290_kDa |
|
| TMTC3 |
transmembrane_and_TPR_repeat-containing_protein_3 |
|
| KITLG |
kit_ligand_isoform_b_precursor |
|
| LOC728084 |
hypothetical_LOC728084 |
|
| LOC100287355 |
|
|
| MRPS6P4 |
|
|
| DUSP6 |
dual_specificity_protein_phosphatase_6_isoform_b |
|
| GALNT4 |
polypeptide_N-acetylgalactosaminyltransferase_4 |
|
| ATP2B1 |
plasma_membrane_calcium-transporting_ATPase_1_isoform_1a |
|
| LOC338758 |
hypothetical_LOC338758 |
|
| MRPL2P1 |
|
|
| LOC100287505 |
|
|
| LOC100507594 |
hypothetical_LOC100507594 |
|
| C12orf12 |
hypothetical_protein_LOC196477 |
|
| EPYC |
epiphycan_precursor |
|
| KERA |
keratocan_precursor |
|
| LUM |
lumican_precursor |
|
| DCN |
decorin_isoform_e_precursor |
|
| BTG1 |
protein_BTG1 |
| 2 |
6568573 |
chr5 |
NT_006576.16 |
21872645-28441218 |
GCNT1P2 |
pro-melanin-concentrating_hormone-like_1 |
|
| LOC100420804 |
|
|
| LOC391771 |
|
|
| PRDM9 |
histone-lysine_N-methyltransferase_PRDM9 |
|
| LOC100130746 |
|
|
| CDH10 |
cadherin-10_isoform_2 |
|
| LOC100422502 |
|
|
| LOC340107 |
hypothetical_LOC340107 |
|
| LOC100506287 |
hypothetical_protein_LOC100506287 |
|
| MSNL1 |
|
|
| - |
tRNA-Lys |
|
| LOC100131678 |
|
|
| CDH9 |
cadherin-9_preproprotein_preproprotein |
| 3 |
6067031 |
chrX |
NT_011651.17 |
16892928-22959959 |
PAICSP7 |
|
|
| CCNB1IP1P3 |
|
|
| MIR548M |
microRNA_548m |
|
| CALM1P1 |
|
|
| LOC100129001 |
|
|
| LOC100128595 |
|
|
| RPS7P13 |
|
|
| LOC100420872 |
|
|
| LOC648927 |
|
|
| RPS29P28 |
|
|
| LOC643486 |
bromodomain,_testis-specific_pseudogene |
|
| LOC100130500 |
|
|
| LOC100422431 |
replication_protein_A_30_kDa_subunit |
|
| LOC100131072 |
|
|
| RPL6P29 |
|
|
| LOC100420955 |
|
|
| EEF1A1P15 |
|
|
| LOC100419995 |
|
|
| LOC100422469 |
X-ray_repair_complementing_defective_repair_pseudogene |
|
| LOC643605 |
|
|
| RPSAP8 |
|
|
| PCDH19 |
protocadherin-19_isoform_c_precursor |
| 4 |
5129370 |
chr8 |
NT_008046.16 |
25355029-30484399 |
EEF1A1P37 |
|
|
| RPL18P7 |
microRNA:hsa-mir-2053 |
|
| LOC100420746 |
|
|
| LOC100506910 |
hypothetical_LOC100506910 |
| 5 |
4897598 |
chr2 |
NT_022135.16 |
12243098-17140696 |
MKI67IP |
MKI67_FHA_domain-interacting_nucleolar_phosphoprotein |
|
| TSN |
translin |
|
| LOC100422580 |
|
|
| LOC100131284 |
|
|
| CNTNAP5 |
contactin-associated_protein-like_5_precursor |
|
| LOC100422417 |
|
|
| LOC150554 |
|
| 6 |
4871759 |
chr4 |
NT_022778.16 |
3170669-8042428 |
RPS15AP17 |
|
|
| RPL21P47 |
|
|
| LOC100289193 |
|
|
| LOC100131441 |
|
|
| LOC644534 |
|
|
| LOC644548 |
|
|
| LOC644578 |
hypothetical_protein_LOC644578 |
|
| TECRL |
trans-2,3-enoyl-CoA_reductase-like |
|
| LOC391657 |
|
|
| RPS6P5 |
|
|
| LOC401134 |
hypothetical_LOC401134 |
|
| LOC100422019 |
|
|
| LOC100507063 |
hypothetical_LOC100507063 |
|
| LOC100144602 |
hypothetical_LOC100144602 |
|
| LOC728048 |
|
|
| MIR1269 |
microRNA:hsa-mir-1269 |
|
| RPS23P3 |
|
| 7 |
4843631 |
chrX |
NT_011651.17 |
2642081-7485712 |
FAM46D |
hypothetical_protein_LOC169966 |
|
| LOC727874 |
|
|
| HK2P1 |
|
|
| LOC100422286 |
bromodomain_and_WD_repeat-containing_protein_3 |
|
| VDAC1P1 |
|
|
| LOC100421037 |
|
|
| HMGN5 |
high_mobility_group_nucleosome-binding_domain-containing_protein_5 |
|
| SH3BGRL |
SH3_domain-binding_glutamic_acid-rich-like_protein |
|
| LOC100422471 |
|
|
| LOC100129843 |
|
|
| RPL22P22 |
|
|
| LOC266683 |
|
|
| POU3F4 |
POU_domain,_class_3,_transcription_factor_4 |
|
| TERF1P4 |
|
|
| CYLC1 |
cylicin-1 |
|
| RPS6KA6 |
ribosomal_protein_S6_kinase_alpha-6 |
|
| MIR548I4 |
microRNA:hsa-mir-548i-4 |
|
| HDX |
highly_divergent_homeobox_isoform_2 |
|
| LOC642869 |
|
|
| UBE2DNL |
ubiquitin-conjugating_enzyme_E2D_N-terminal_like_(pseudogene) |
| 8 |
4480536 |
chr11 |
NT_033899.8 |
5780444-10260980 |
BIRC2 |
baculoviral_IAP_repeat-containing_protein_2 |
|
| TMEM123 |
porimin_precursor |
|
| LOC727869 |
hypothetical_LOC727869 |
|
| MMP7 |
matrilysin_preproprotein_preproprotein |
|
| MMP20 |
matrix_metalloproteinase-20_preproprotein_preproprotein |
|
| MMP27 |
matrix_metalloproteinase-27_precursor |
|
| MMP8 |
neutrophil_collagenase_preproprotein_preproprotein |
|
| LOC100421658 |
|
|
| MMP10 |
stromelysin-2_preproprotein_preproprotein |
|
| CSNK1A1P2 |
interstitial_collagenase_isoform_2 |
|
| MMP3 |
stromelysin-1_preproprotein_preproprotein |
|
| MMP12 |
macrophage_metalloelastase_preproprotein_preproprotein |
|
| LOC100288111 |
|
|
| MMP13 |
collagenase_3_preproprotein_preproprotein |
|
| RPL21P96 |
|
|
| DCUN1D5 |
DCN1-like_protein_5 |
|
| LOC100506721 |
cytoplasmic_dynein_2_heavy_chain_1_isoform_2 |
|
| LOC100190922 |
|
|
| DDI1 |
protein_DDI1_homolog_1 |
|
| LOC100506742 |
inactive_caspase-12-like_isoform_2 |
|
| LOC643733 |
caspase_4,_apoptosis-related_cysteine_peptidase_pseudogene,_transcript_variant_2 |
|
| CASP4 |
caspase-4_isoform_gamma_precursor |
|
| CASP5 |
caspase-5_isoform_f_precursor |
|
| CASP1 |
caspase-1_isoform_gamma_precursor |
|
| CARD16 |
caspase_recruitment_domain-containing_protein_16_isoform_2 |
|
| LOC440067 |
|
|
| CARD17 |
caspase_recruitment_domain-containing_protein_17 |
|
| CARD18 |
caspase_recruitment_domain-containing_protein_18 |
|
| OR2AL1P |
|
|
| HSPD1P13 |
glutamate_receptor_4_isoform_3_precursor |
|
| KIAA1826 |
hypothetical_protein_LOC84437 |
|
| KBTBD3 |
kelch_repeat_and_BTB_domain-containing_protein_3 |
|
| AASDHPPT |
L-aminoadipate-semialdehyde_dehydrogenase-phosphopantetheinyl_transferase |
|
| LOC643855 |
|
|
| LOC100422300 |
guanylate_cyclase_soluble_subunit_alpha-2 |
|