| Enzyme: | XorKI | Longest uncut segments |
| Specificity: | CGATCG | Repeats in uncut segments |
| Number of sites: | 12145 | Genes in uncut segments |
| Mean distance between sites: | 235598 base pairs | |
| Standard deviation: | 295546 base pairs | |
| Site density | 4.2 per megabase | Help |
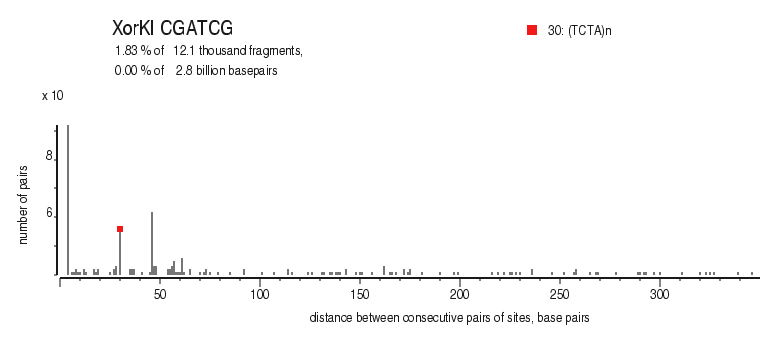
| Distribution of closely spaced sites |
|---|
 |
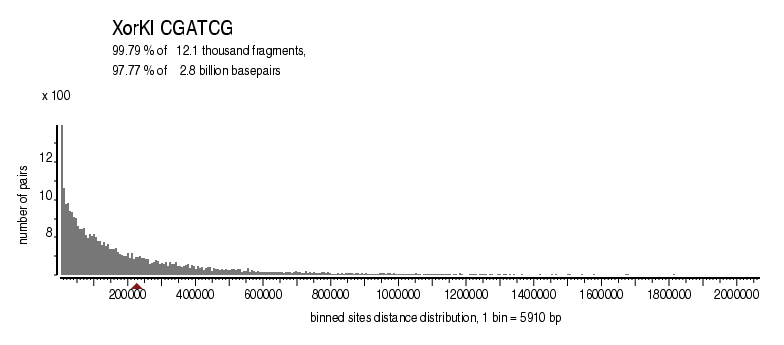
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 3299736 | chr13 | NT_024524.14 | 37270597-40570333 | 50.65 % in | 5069 repeats | 3.71 % in | 12 genes |
| 2 | 3295865 | chr1 | NT_032977.9 | 73383882-76679747 | 56.68 % in | 5078 repeats | 13.34 % in | 15 genes |
| 3 | 2854362 | chr5 | NT_034772.6 | 11212600-14066962 | 50.48 % in | 4505 repeats | 0.08 % in | 2 genes |
| 4 | 2779316 | chrX | NT_011651.17 | 2827759-5607075 | 76.99 % in | 3908 repeats | 16.30 % in | 12 genes |
| 5 | 2754037 | chr12 | NT_029419.12 | 44486218-47240255 | 53.74 % in | 4298 repeats | 20.96 % in | 6 genes |
| 6 | 2735689 | chrX | NT_011651.17 | 16532703-19268392 | 69.35 % in | 4298 repeats | 1.62 % in | 14 genes |
| 7 | 2497192 | chr1 | NT_004487.19 | 45824392-48321584 | 52.56 % in | 3957 repeats | 20.96 % in | 5 genes |
| 8 | 2384773 | chr9 | NT_008470.19 | 12092440-14477213 | 50.86 % in | 3870 repeats | 7.10 % in | 14 genes |
| 9 | 2378724 | chr5 | NT_006576.16 | 26132420-28511144 | 52.48 % in | 3830 repeats | 0.00 % in | 0 genes |
| 10 | 2352485 | chr7 | NT_007933.15 | 24087989-26440474 | 50.45 % in | 3709 repeats | 0.00 % in | 0 genes |
| 11 | 2351983 | chr6 | NT_007299.13 | 19072530-21424513 | 54.65 % in | 3580 repeats | 0.00 % in | 0 genes |
| 12 | 2329985 | chr6 | NT_025741.15 | 26168783-28498768 | 48.31 % in | 3685 repeats | 0.00 % in | 0 genes |
| 13 | 2320818 | chr3 | NT_005612.16 | 51021294-53342112 | 54.05 % in | 3669 repeats | 0.00 % in | 0 genes |
| 14 | 2284965 | chr7 | NT_007933.15 | 17820658-20105623 | 40.77 % in | 3533 repeats | 0.00 % in | 0 genes |
| 15 | 2277338 | chr2 | NT_022135.16 | 29407211-31684549 | 45.69 % in | 3591 repeats | 0.00 % in | 0 genes |
| 16 | 2271842 | chr13 | NT_024524.14 | 46923008-49194850 | 40.90 % in | 3259 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 3299736 | chr13 | NT_024524.14 | 37270597-40570333 | 5069 | 554 | AT_rich (674) | AluSx (148) | L2a (144) | |
| 2 | 3295865 | chr1 | NT_032977.9 | 73383882-76679747 | 5078 | 521 | AT_rich (683) | L2a (170) | (TA)n (138) | |
| 3 | 2854362 | chr5 | NT_034772.6 | 11212600-14066962 | 4505 | 558 | AT_rich (567) | AluSx (131) | L2a (130) | |
| 4 | 2779316 | chrX | NT_011651.17 | 2827759-5607075 | 3908 | 480 | AT_rich (243) | AluSx (113) | (TA)n (86) | |
| 5 | 2754037 | chr12 | NT_029419.12 | 44486218-47240255 | 4298 | 526 | AT_rich (399) | L2a (170) | MIRb (152) | |
| 6 | 2735689 | chrX | NT_011651.17 | 16532703-19268392 | 4298 | 531 | AT_rich (279) | MIR (108) | AluSx (108) | |
| 7 | 2497192 | chr1 | NT_004487.19 | 45824392-48321584 | 3957 | 487 | AT_rich (535) | L2a (122) | AluSx (119) | |
| 8 | 2384773 | chr9 | NT_008470.19 | 12092440-14477213 | 3870 | 479 | AT_rich (237) | AluSx (174) | AluY (165) | |
| 9 | 2378724 | chr5 | NT_006576.16 | 26132420-28511144 | 3830 | 492 | AT_rich (469) | (TA)n (121) | AluSx (117) | |
| 10 | 2352485 | chr7 | NT_007933.15 | 24087989-26440474 | 3709 | 468 | AT_rich (291) | MIRb (146) | L2a (146) | |
| 11 | 2351983 | chr6 | NT_007299.13 | 19072530-21424513 | 3580 | 455 | AT_rich (311) | MIRb (152) | AluSx (124) | |
| 12 | 2329985 | chr6 | NT_025741.15 | 26168783-28498768 | 3685 | 489 | AT_rich (385) | L2a (151) | MIRb (148) | |
| 13 | 2320818 | chr3 | NT_005612.16 | 51021294-53342112 | 3669 | 519 | AT_rich (416) | L2a (116) | MIR (106) | |
| 14 | 2284965 | chr7 | NT_007933.15 | 17820658-20105623 | 3533 | 466 | AT_rich (399) | MIR (147) | AluY (141) | |
| 15 | 2277338 | chr2 | NT_022135.16 | 29407211-31684549 | 3591 | 492 | AT_rich (413) | MIR (143) | MIRb (142) | |
| 16 | 2271842 | chr13 | NT_024524.14 | 46923008-49194850 | 3259 | 460 | AT_rich (465) | MIRb (133) | MIR (110) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 3299736 | chr13 | NT_024524.14 | 37270597-40570333 | HNF4GP1 | |
| PRR20A | proline-rich_protein_20A | |||||
| PRR20B | proline-rich_protein_20B | |||||
| PRR20C | proline-rich_protein_20C | |||||
| PRR20D | proline-rich_protein_20D | |||||
| PRR20E | proline-rich_protein_20E | |||||
| SLC25A5P4 | ||||||
| RPL31P53 | ||||||
| LOC100129744 | hypothetical_protein_LOC100129744 | |||||
| TRNAE39P | ||||||
| LOC341689 | ||||||
| LOC100129308 | ||||||
| 2 | 3295865 | chr1 | NT_032977.9 | 73383882-76679747 | LOC100421046 | collagen_alpha-1(XI)_chain_isoform_E_preproprotein |
| RNPC3 | RNA-binding_protein_40 | |||||
| AMY2B | alpha-amylase_2B_precursor | |||||
| AMY2A | pancreatic_alpha-amylase_precursor | |||||
| AMY1A | alpha-amylase_1_precursor | |||||
| AMY1B | alpha-amylase_1_precursor | |||||
| AMYP1 | ||||||
| AMY1C | alpha-amylase_1_precursor | |||||
| LOC100131348 | ||||||
| LOC100129138 | THAP_domain_containing,_apoptosis_associated_protein_3_pseudogene | |||||
| FTLP17 | ||||||
| CDK4PS | ||||||
| LOC100499497 | ||||||
| LOC401957 | ||||||
| LOC126987 | ||||||
| 3 | 2854362 | chr5 | NT_034772.6 | 11212600-14066962 | RAB9BP1 | RAB9B,_member_RAS_oncogene_family_pseudogene_1 |
| LOC345571 | ||||||
| 4 | 2779316 | chrX | NT_011651.17 | 2827759-5607075 | FAM46D | hypothetical_protein_LOC169966 |
| LOC727874 | ||||||
| HK2P1 | ||||||
| LOC100422286 | bromodomain_and_WD_repeat-containing_protein_3 | |||||
| VDAC1P1 | ||||||
| LOC100421037 | ||||||
| HMGN5 | high_mobility_group_nucleosome-binding_domain-containing_protein_5 | |||||
| SH3BGRL | SH3_domain-binding_glutamic_acid-rich-like_protein | |||||
| LOC100422471 | ||||||
| LOC100129843 | ||||||
| RPL22P22 | ||||||
| LOC266683 | ||||||
| 5 | 2754037 | chr12 | NT_029419.12 | 44486218-47240255 | CCDC59 | thyroid_transcription_factor_1-associated_protein_26 |
| LOC100421061 | hypothetical_protein_LOC84190 | |||||
| LOC100418732 | ||||||
| TMTC2 | transmembrane_and_TPR_repeat-containing_protein_2 | |||||
| RPL6P25 | ||||||
| LOC100128335 | ||||||
| 6 | 2735689 | chrX | NT_011651.17 | 16532703-19268392 | LOC643371 | |
| PAICSP7 | ||||||
| CCNB1IP1P3 | ||||||
| MIR548M | microRNA_548m | |||||
| CALM1P1 | ||||||
| LOC100129001 | ||||||
| LOC100128595 | ||||||
| RPS7P13 | ||||||
| LOC100420872 | ||||||
| LOC648927 | ||||||
| RPS29P28 | ||||||
| LOC643486 | bromodomain,_testis-specific_pseudogene | |||||
| LOC100130500 | ||||||
| LOC100422431 | replication_protein_A_30_kDa_subunit | |||||
| 7 | 2497192 | chr1 | NT_004487.19 | 45824392-48321584 | KCNT2 | potassium_channel_subfamily_T_member_2 |
| CFH | complement_factor_H_isoform_b_precursor | |||||
| CFHR3 | complement_factor_H-related_protein_3_isoform_2_precursor | |||||
| CFHR1 | complement_factor_H-related_protein_1_precursor | |||||
| LOC100289145 | ||||||
| 8 | 2384773 | chr9 | NT_008470.19 | 12092440-14477213 | LOC100287067 | |
| RPS19P6 | ||||||
| RPS20P25 | ||||||
| TLE1 | transducin-like_enhancer_protein_1 | |||||
| FAM75D5 | family_with_sequence_similarity_75,_member_D5 | |||||
| FAM75D4 | hypothetical_protein_LOC389761 | |||||
| FAM75D3 | hypothetical_protein_LOC389762 | |||||
| FAM75D2P | ||||||
| FAM75D1 | hypothetical_protein_LOC389763 | |||||
| FAM75B | family_with_sequence_similarity_75,_member_B | |||||
| LOC401533 | ||||||
| RPS2P34 | ||||||
| LOC100420670 | ||||||
| LOC442427 | ||||||