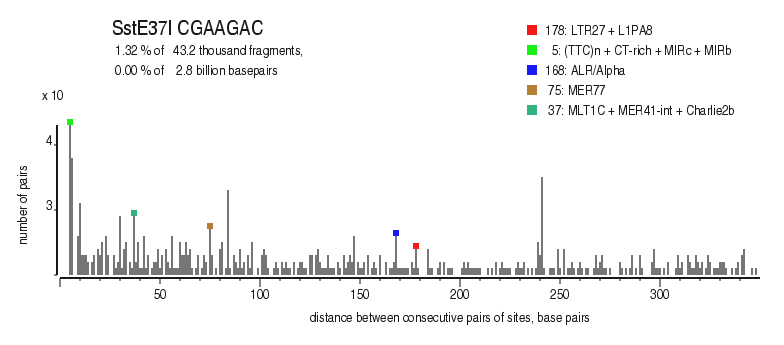
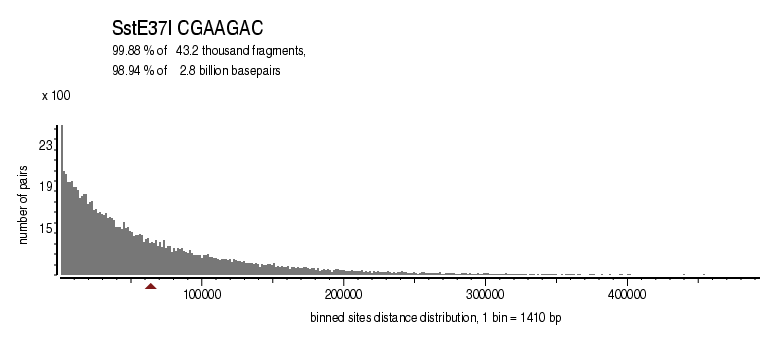
| Enzyme: | SstE37I | Longest uncut segments |
| Specificity: | CGAAGAC | Repeats in uncut segments |
| Number of sites: | 43249 | Genes in uncut segments |
| Mean distance between sites: | 66159 base pairs | |
| Standard deviation: | 70539 base pairs | |
| Site density | 15.1 per megabase | Help |
| Distribution of closely spaced sites |
|---|
 |
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 957253 | chr1 | NT_032977.9 | 40714432-41671685 | 54.86 % in | 1721 repeats | 37.25 % in | 8 genes |
| 2 | 884803 | chr6 | NT_007592.15 | 50091879-50976682 | 42.96 % in | 1300 repeats | 10.42 % in | 6 genes |
| 3 | 804405 | chr2 | NT_005403.17 | 64966820-65771225 | 48.71 % in | 1294 repeats | 84.79 % in | 2 genes |
| 4 | 689740 | chr13 | NT_024524.14 | 41491434-42181174 | 46.84 % in | 1218 repeats | 59.58 % in | 3 genes |
| 5 | 680937 | chr14 | NT_026437.12 | 27939357-28620294 | 46.07 % in | 1057 repeats | 47.54 % in | 3 genes |
| 6 | 676324 | chr13 | NT_024524.14 | 42786908-43463232 | 55.61 % in | 1073 repeats | 0.86 % in | 1 genes |
| 7 | 672874 | chr6 | NT_007299.13 | 4871496-5544370 | 57.66 % in | 1039 repeats | 0.71 % in | 2 genes |
| 8 | 671309 | chr19 | NT_011295.11 | 12463149-13134458 | 70.92 % in | 1448 repeats | 45.51 % in | 15 genes |
| 9 | 645192 | chr4 | NT_016354.19 | 24006035-24651227 | 56.52 % in | 1252 repeats | 0.00 % in | 0 genes |
| 10 | 644275 | chr6 | NT_007299.13 | 19744367-20388642 | 57.74 % in | 985 repeats | 0.00 % in | 0 genes |
| 11 | 620016 | chr7 | NT_007933.15 | 20295536-20915552 | 41.15 % in | 923 repeats | 0.00 % in | 0 genes |
| 12 | 618902 | chr3 | NT_005612.16 | 86362993-86981895 | 55.94 % in | 1056 repeats | 0.00 % in | 0 genes |
| 13 | 615887 | chr12 | NT_009775.17 | 3387152-4003039 | 59.93 % in | 1695 repeats | 0.00 % in | 0 genes |
| 14 | 610852 | chr4 | NT_022778.16 | 10313239-10924091 | 58.35 % in | 905 repeats | 0.00 % in | 0 genes |
| 15 | 608698 | chr13 | NT_024524.14 | 47616687-48225385 | 36.66 % in | 863 repeats | 0.00 % in | 0 genes |
| 16 | 606591 | chr4 | NT_006316.16 | 17801412-18408003 | 52.12 % in | 1088 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 957253 | chr1 | NT_032977.9 | 40714432-41671685 | 1721 | 339 | AT_rich (101) | AluSx (101) | MIRb (94) | |
| 2 | 884803 | chr6 | NT_007592.15 | 50091879-50976682 | 1300 | 274 | AT_rich (118) | MIR (61) | MIRb (59) | |
| 3 | 804405 | chr2 | NT_005403.17 | 64966820-65771225 | 1294 | 298 | AT_rich (108) | L2a (55) | MIR (49) | |
| 4 | 689740 | chr13 | NT_024524.14 | 41491434-42181174 | 1218 | 286 | AT_rich (87) | AluSx (70) | L2a (53) | |
| 5 | 680937 | chr14 | NT_026437.12 | 27939357-28620294 | 1057 | 268 | AT_rich (142) | L2a (42) | MIR (41) | |
| 6 | 676324 | chr13 | NT_024524.14 | 42786908-43463232 | 1073 | 290 | AT_rich (134) | L2a (33) | L2 (31) | |
| 7 | 672874 | chr6 | NT_007299.13 | 4871496-5544370 | 1039 | 282 | AT_rich (133) | (TA)n (30) | L2a (30) | |
| 8 | 671309 | chr19 | NT_011295.11 | 12463149-13134458 | 1448 | 183 | AluSx (142) | AluSg (90) | AluSp (80) | |
| 9 | 645192 | chr4 | NT_016354.19 | 24006035-24651227 | 1252 | 292 | AluSx (82) | AT_rich (63) | AluJo (50) | |
| 10 | 644275 | chr6 | NT_007299.13 | 19744367-20388642 | 985 | 237 | AT_rich (100) | (TA)n (30) | MIRb (27) | |
| 11 | 620016 | chr7 | NT_007933.15 | 20295536-20915552 | 923 | 251 | AT_rich (124) | MIR (33) | MIRb (31) | |
| 12 | 618902 | chr3 | NT_005612.16 | 86362993-86981895 | 1056 | 254 | AT_rich (73) | MIRb (54) | L2a (41) | |
| 13 | 615887 | chr12 | NT_009775.17 | 3387152-4003039 | 1695 | 261 | AluSx (155) | MIR (130) | MIRb (116) | |
| 14 | 610852 | chr4 | NT_022778.16 | 10313239-10924091 | 905 | 228 | AT_rich (112) | MIR (25) | L1ME3A (20) | |
| 15 | 608698 | chr13 | NT_024524.14 | 47616687-48225385 | 863 | 234 | AT_rich (118) | L2a (35) | MIRb (33) | |
| 16 | 606591 | chr4 | NT_006316.16 | 17801412-18408003 | 1088 | 255 | AT_rich (69) | L2a (59) | L2c (56) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 957253 | chr1 | NT_032977.9 | 40714432-41671685 | ANKRD13C | ankyrin_repeat_domain-containing_protein_13C |
| HHLA3 | HERV-H_LTR-associating_protein_3_isoform_3 | |||||
| CTH | cystathionine_gamma-lyase_isoform_3 | |||||
| LOC391048 | ||||||
| LOC100131616 | ||||||
| PTGER3 | prostaglandin_E2_receptor_EP3_subtype_isoform_5 | |||||
| LOC100132618 | hypothetical_LOC100132618 | |||||
| MIR186 | microRNA:hsa-mir-186 | |||||
| 2 | 884803 | chr6 | NT_007592.15 | 50091879-50976682 | TFAP2D | transcription_factor_AP-2-delta |
| TFAP2B | transcription_factor_AP-2-beta | |||||
| RPS17P5 | ||||||
| LOC100418898 | ||||||
| FTH1P5 | ||||||
| LOC100421020 | ||||||
| 3 | 804405 | chr2 | NT_005403.17 | 64966820-65771225 | RPL5P8 | sperm-associated_antigen_16_protein_isoform_2 |
| VWC2L | von_Willebrand_factor_C_domain-containing_protein_2-like_precursor | |||||
| 4 | 689740 | chr13 | NT_024524.14 | 41491434-42181174 | DIAPH3 | protein_diaphanous_homolog_3_isoform_b |
| LOC440142 | ||||||
| TDRD3 | tudor_domain-containing_protein_3_isoform_2 | |||||
| 5 | 680937 | chr14 | NT_026437.12 | 27939357-28620294 | LOC100506412 | hypothetical_LOC100506412 |
| RPL10L | 60S_ribosomal_protein_L10-like | |||||
| RPS15AP3 | MAM_domain-containing_glycosylphosphatidylinositol_anchor_protein_2_isoform_2 | |||||
| 6 | 676324 | chr13 | NT_024524.14 | 42786908-43463232 | PCDH20 | protocadherin-20 |
| 7 | 672874 | chr6 | NT_007299.13 | 4871496-5544370 | NUFIP1P | |
| LOC100420940 | ||||||
| 8 | 671309 | chr19 | NT_011295.11 | 12463149-13134458 | ZNF430 | zinc_finger_protein_430_isoform_2 |
| VN1R80P | ||||||
| VN1R81P | zinc_finger_protein_714 | |||||
| RPL7AP10 | zinc_finger_protein_431 | |||||
| VN1R82P | ||||||
| RPL36AP51 | ||||||
| VN1R83P | ||||||
| LOC100421707 | zinc_finger_protein_708 | |||||
| LOC100422298 | zinc_finger_protein_738 | |||||
| ZNF493 | zinc_finger_protein_493_isoform_1 | |||||
| LOC400680 | hypothetical_LOC400680 | |||||
| LOC100421699 | zinc_finger_protein_429 | |||||
| LOC100505594 | hypothetical_LOC100505594 | |||||
| LOC400682 | zinc_finger_protein_100 | |||||
| LOC100418832 | ||||||