| Enzyme: | SdaI | Longest uncut segments |
| Specificity: | CCTGCAGG | Repeats in uncut segments |
| Number of sites: | 76937 | Genes in uncut segments |
| Mean distance between sites: | 37190 base pairs | |
| Standard deviation: | 53243 base pairs | |
| Site density | 26.9 per megabase | Help |
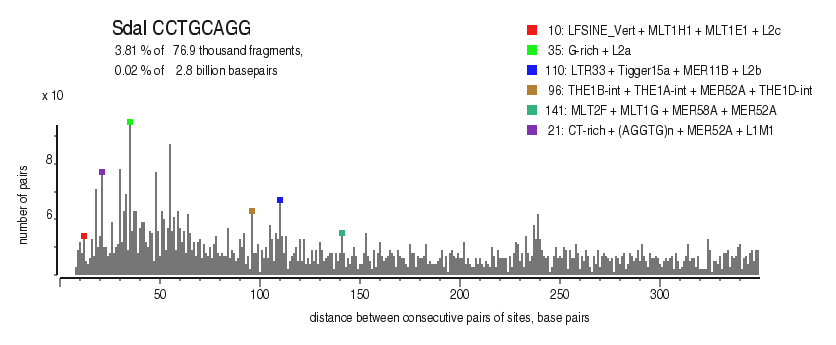
| Distribution of closely spaced sites |
|---|
 |
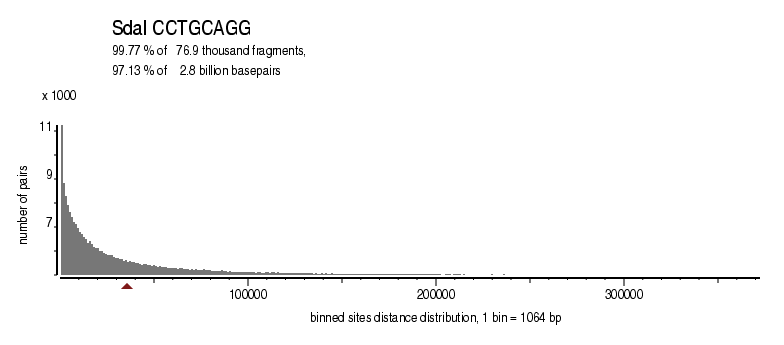
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 1056072 | chr3 | NT_005612.16 | 70706663-71762735 | 49.73 % in | 1758 repeats | 10.37 % in | 2 genes |
| 2 | 865730 | chr13 | NT_024524.14 | 64294729-65160459 | 52.13 % in | 1375 repeats | 0.20 % in | 1 genes |
| 3 | 810071 | chr9 | NT_008413.18 | 20746649-21556720 | 48.65 % in | 1169 repeats | 48.67 % in | 27 genes |
| 4 | 746365 | chr6 | NT_007299.13 | 5766099-6512464 | 56.49 % in | 1191 repeats | 0.12 % in | 1 genes |
| 5 | 744422 | chr4 | NT_016354.19 | 87405585-88150007 | 49.63 % in | 1260 repeats | 30.64 % in | 2 genes |
| 6 | 732510 | chr13 | NT_024524.14 | 52879572-53612082 | 34.23 % in | 1078 repeats | 58.66 % in | 2 genes |
| 7 | 706626 | chr3 | NT_005612.16 | 50812302-51518928 | 52.53 % in | 1163 repeats | 0.00 % in | 0 genes |
| 8 | 668417 | chr21 | NT_011512.11 | 9034969-9703386 | 56.38 % in | 1158 repeats | 0.69 % in | 2 genes |
| 9 | 661694 | chr13 | NT_009952.14 | 2470353-3132047 | 52.91 % in | 1083 repeats | 0.00 % in | 0 genes |
| 10 | 660942 | chr2 | NT_022135.16 | 31821990-32482932 | 35.19 % in | 967 repeats | 0.00 % in | 0 genes |
| 11 | 656771 | chr5 | NT_006576.16 | 22235429-22892200 | 41.96 % in | 1044 repeats | 0.00 % in | 0 genes |
| 12 | 637271 | chr1 | NT_004487.19 | 25603637-26240908 | 59.84 % in | 1101 repeats | 0.00 % in | 0 genes |
| 13 | 636974 | chr5 | NT_006576.16 | 29023040-29660014 | 55.88 % in | 1030 repeats | 0.00 % in | 0 genes |
| 14 | 632482 | chr8 | NT_008046.16 | 7025068-7657550 | 44.96 % in | 989 repeats | 0.00 % in | 0 genes |
| 15 | 629881 | chr18 | NT_025028.14 | 14229292-14859173 | 41.67 % in | 1021 repeats | 0.00 % in | 0 genes |
| 16 | 622614 | chr12 | NT_029419.12 | 52723324-53345938 | 52.68 % in | 1070 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 1056072 | chr3 | NT_005612.16 | 70706663-71762735 | 1758 | 339 | AT_rich (233) | L2a (57) | MIRb (51) | |
| 2 | 865730 | chr13 | NT_024524.14 | 64294729-65160459 | 1375 | 322 | AT_rich (176) | (TA)n (40) | AluSx (35) | |
| 3 | 810071 | chr9 | NT_008413.18 | 20746649-21556720 | 1169 | 260 | AT_rich (111) | MIRb (57) | AluSx (45) | |
| 4 | 746365 | chr6 | NT_007299.13 | 5766099-6512464 | 1191 | 278 | AT_rich (169) | L2a (44) | (TA)n (43) | |
| 5 | 744422 | chr4 | NT_016354.19 | 87405585-88150007 | 1260 | 307 | AT_rich (133) | L2a (49) | AluY (45) | |
| 6 | 732510 | chr13 | NT_024524.14 | 52879572-53612082 | 1078 | 235 | AT_rich (154) | MIR (53) | MIRb (47) | |
| 7 | 706626 | chr3 | NT_005612.16 | 50812302-51518928 | 1163 | 288 | AT_rich (143) | L2a (39) | MIRb (33) | |
| 8 | 668417 | chr21 | NT_011512.11 | 9034969-9703386 | 1158 | 295 | AT_rich (136) | (TA)n (35) | AluY (32) | |
| 9 | 661694 | chr13 | NT_009952.14 | 2470353-3132047 | 1083 | 268 | AT_rich (143) | AluSx (37) | L2a (28) | |
| 10 | 660942 | chr2 | NT_022135.16 | 31821990-32482932 | 967 | 232 | AT_rich (127) | MIRb (47) | L2c (38) | |
| 11 | 656771 | chr5 | NT_006576.16 | 22235429-22892200 | 1044 | 243 | AT_rich (130) | L2a (48) | AluY (37) | |
| 12 | 637271 | chr1 | NT_004487.19 | 25603637-26240908 | 1101 | 239 | AT_rich (81) | AluSx (64) | MIR (38) | |
| 13 | 636974 | chr5 | NT_006576.16 | 29023040-29660014 | 1030 | 264 | AT_rich (117) | L1M5 (31) | AluSx (30) | |
| 14 | 632482 | chr8 | NT_008046.16 | 7025068-7657550 | 989 | 256 | AT_rich (92) | MIRb (51) | L2c (49) | |
| 15 | 629881 | chr18 | NT_025028.14 | 14229292-14859173 | 1021 | 249 | AT_rich (136) | MIRb (31) | L2a (31) | |
| 16 | 622614 | chr12 | NT_029419.12 | 52723324-53345938 | 1070 | 267 | AT_rich (94) | L2a (45) | L2c (34) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 1056072 | chr3 | NT_005612.16 | 70706663-71762735 | SI | sucrase-isomaltase,_intestinal |
| SLITRK3 | SLIT_and_NTRK-like_protein_3_precursor | |||||
| 2 | 865730 | chr13 | NT_024524.14 | 64294729-65160459 | LOC647317 | |
| 3 | 810071 | chr9 | NT_008413.18 | 20746649-21556720 | MIR491 | microRNA:hsa-mir-491 |
| PTPLAD2 | 3-hydroxyacyl-CoA_dehydratase_4 | |||||
| IFNNP1 | ||||||
| IFNB1 | interferon_beta_precursor | |||||
| IFNW1 | interferon_omega-1_precursor | |||||
| IFNA21 | interferon_alpha-21_precursor | |||||
| IFNWP15 | ||||||
| IFNA4 | interferon_alpha-4_precursor | |||||
| IFNWP9 | ||||||
| IFNA7 | interferon_alpha-7_precursor | |||||
| IFNA10 | interferon_alpha-10_precursor | |||||
| IFNWP18 | ||||||
| IFNA16 | interferon_alpha-16_precursor | |||||
| IFNA17 | interferon_alpha-17_precursor | |||||
| IFNWP5 | ||||||
| IFNA14 | interferon_alpha-14_precursor | |||||
| IFNA22P | ||||||
| IFNA5 | interferon_alpha-5_precursor | |||||
| KLHL9 | kelch-like_protein_9 | |||||
| IFNA6 | interferon_alpha-6_precursor | |||||
| IFNA13 | interferon_alpha-1/13_precursor | |||||
| IFNA2 | interferon_alpha-2_precursor | |||||
| IFNWP12 | ||||||
| IFNA8 | interferon_alpha-8_precursor | |||||
| IFNWP2 | ||||||
| IFNA1 | interferon_alpha-1/13_precursor | |||||
| MIR31 | microRNA:hsa-mir-31 | |||||
| 4 | 746365 | chr6 | NT_007299.13 | 5766099-6512464 | LOC100420211 | |
| 5 | 744422 | chr4 | NT_016354.19 | 87405585-88150007 | FSTL5 | follistatin-related_protein_5_isoform_c |
| LOC100131135 | ||||||
| 6 | 732510 | chr13 | NT_024524.14 | 52879572-53612082 | RPL35AP31 | dachshund_homolog_1_isoform_c |
| RPS10P21 | ||||||
| 8 | 668417 | chr21 | NT_011512.11 | 9034969-9703386 | LOC100420035 | |
| MAPK6PS2 | ||||||