| Enzyme: | BbvCIB | Longest uncut segments |
| Specificity: | CCTCAGC | Repeats in uncut segments |
| Number of sites: | 1389763 | Genes in uncut segments |
| Mean distance between sites: | 2058 base pairs | |
| Standard deviation: | 2797 base pairs | |
| Site density | 485.7 per megabase | Help |
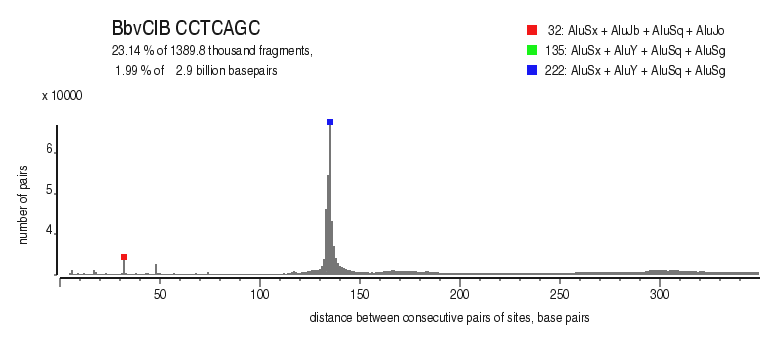
| Distribution of closely spaced sites |
|---|
 |
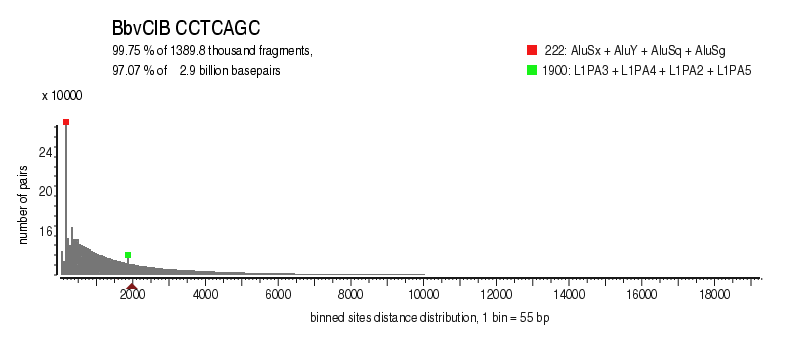
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 494168 | chr15 | NT_037852.6 | 1397320-1891488 | 0.76 % in | 18 repeats | 0.00 % in | 0 genes |
| 2 | 404502 | chr6 | NT_167244.1 | 2359778-2764280 | 0.13 % in | 3 repeats | 0.00 % in | 0 genes |
| 3 | 269129 | chrY | NT_011875.12 | 8417470-8686599 | 81.29 % in | 21 repeats | 0.00 % in | 0 genes |
| 4 | 224805 | chr7 | NT_023603.5 | 14949-239754 | 98.02 % in | 30 repeats | 0.00 % in | 0 genes |
| 5 | 212277 | chr6 | NT_167244.1 | 4386491-4598768 | 2.05 % in | 13 repeats | 0.00 % in | 0 genes |
| 6 | 184684 | chr6 | NT_167244.1 | 3786305-3970989 | 0.68 % in | 9 repeats | 0.64 % in | 1 genes |
| 7 | 183442 | chr4 | NT_006316.16 | 378663-562105 | 8.08 % in | 76 repeats | 19.63 % in | 23 genes |
| 8 | 178096 | chr6 | NT_167244.1 | 3177289-3355385 | 0.73 % in | 15 repeats | 1.65 % in | 1 genes |
| 9 | 174424 | chr6 | NT_167247.1 | 4420725-4595149 | 0.72 % in | 6 repeats | 0.00 % in | 0 genes |
| 10 | 170163 | chr6 | NT_167248.1 | 512068-682231 | 6.09 % in | 7 repeats | 0.00 % in | 0 genes |
| 11 | 166992 | chr6 | NT_167247.1 | 1560534-1727526 | 0.92 % in | 7 repeats | 0.00 % in | 0 genes |
| 12 | 166638 | chr6 | NT_167249.1 | 2137129-2303767 | 0.80 % in | 7 repeats | 0.00 % in | 0 genes |
| 13 | 153360 | chr9 | NT_008470.19 | 21691002-21844362 | 0.98 % in | 7 repeats | 0.00 % in | 0 genes |
| 14 | 143652 | chr6 | NT_167244.1 | 2894137-3037789 | 0.35 % in | 5 repeats | 0.00 % in | 0 genes |
| 15 | 139663 | chr9 | NT_078070.3 | 949560-1089223 | 60.05 % in | 39 repeats | 0.00 % in | 0 genes |
| 16 | 131245 | chr10 | NT_008705.16 | 38707414-38838659 | 27.39 % in | 232 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 494168 | chr15 | NT_037852.6 | 1397320-1891488 | 18 | 15 | L2a (3) | L1M5 (2) | U2 (1) | |
| 2 | 404502 | chr6 | NT_167244.1 | 2359778-2764280 | 3 | 3 | L1MEg (1) | AluY (1) | AluSp (1) | |
| 3 | 269129 | chrY | NT_011875.12 | 8417470-8686599 | 21 | 4 | LTR12B (17) | LTR12D (2) | L1PA7 (1) | |
| 4 | 224805 | chr7 | NT_023603.5 | 14949-239754 | 30 | 18 | ALR/Alpha (6) | L1PA2 (4) | AT_rich (3) | |
| 5 | 212277 | chr6 | NT_167244.1 | 4386491-4598768 | 13 | 9 | MER57-int (3) | AluSx (3) | (TTTTA)n (1) | |
| 6 | 184684 | chr6 | NT_167244.1 | 3786305-3970989 | 9 | 8 | AT_rich (2) | MLT1H-int (1) | MIR (1) | |
| 7 | 183442 | chr4 | NT_006316.16 | 378663-562105 | 76 | 20 | (CA)n (48) | L1M4 (7) | L1PA10 (2) | |
| 8 | 178096 | chr6 | NT_167244.1 | 3177289-3355385 | 15 | 11 | GC_rich (3) | LTR23 (2) | AluSx (2) | |
| 9 | 174424 | chr6 | NT_167247.1 | 4420725-4595149 | 6 | 6 | MIR (1) | MER11A (1) | L2b (1) | |
| 10 | 170163 | chr6 | NT_167248.1 | 512068-682231 | 7 | 7 | LTR7 (1) | L1PREC2 (1) | L1PA7 (1) | |
| 11 | 166992 | chr6 | NT_167247.1 | 1560534-1727526 | 7 | 7 | Tigger7 (1) | MIRc (1) | MIR (1) | |
| 12 | 166638 | chr6 | NT_167249.1 | 2137129-2303767 | 7 | 5 | L1MB8 (2) | AluSx (2) | L1MC4a (1) | |
| 13 | 153360 | chr9 | NT_008470.19 | 21691002-21844362 | 7 | 6 | LTR67B (2) | MSTA (1) | MIR3 (1) | |
| 14 | 143652 | chr6 | NT_167244.1 | 2894137-3037789 | 5 | 5 | L1MC5 (1) | AluY (1) | AluSg1 (1) | |
| 15 | 139663 | chr9 | NT_078070.3 | 949560-1089223 | 39 | 20 | ERVL-E-int (5) | (TAA)n (3) | L1PA3 (3) | |
| 16 | 131245 | chr10 | NT_008705.16 | 38707414-38838659 | 232 | 43 | GA-rich (24) | (GAATG)n (22) | (AAATG)n (22) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 6 | 184684 | chr6 | NT_167244.1 | 3786305-3970989 | HLA-DRB3 | major_histocompatibility_complex,_class_II,_DR_beta_3_precursor |
| 7 | 183442 | chr4 | NT_006316.16 | 378663-562105 | LOC100287144 | ubiquitin_carboxyl-terminal_hydrolase_17-like |
| LOC100287178 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| LOC100287205 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| LOC100287238 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| LOC100287270 | ||||||
| LOC100288520 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| LOC100287302 | ||||||
| LOC100287327 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| LOC100287364 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| LOC100287404 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| LOC100287441 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| LOC100287478 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| LOC100287513 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| LOC728369 | ubiquitin_carboxyl-terminal_hydrolase_17 | |||||
| LOC728373 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| LOC728379 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| USP17L5 | ubiquitin_specific_peptidase_17-like_5 | |||||
| LOC728393 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| LOC728400 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| LOC728405 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| USP17 | ubiquitin_carboxyl-terminal_hydrolase_17 | |||||
| LOC728419 | ubiquitin_carboxyl-terminal_hydrolase_17-like | |||||
| USP17L6P | ubiquitin_specific_peptidase_17-like_6_(pseudogene) | |||||
| 8 | 178096 | chr6 | NT_167244.1 | 3177289-3355385 | EHMT2 | histone-lysine_N-methyltransferase,_H3_lysine-9_specific_3_isoform_b |