| Enzyme: | SenpCI | Longest uncut segments |
| Specificity: | CCGCGG | Repeats in uncut segments |
| Number of sites: | 65238 | Genes in uncut segments |
| Mean distance between sites: | 43860 base pairs | |
| Standard deviation: | 107870 base pairs | |
| Site density | 22.8 per megabase | Help |
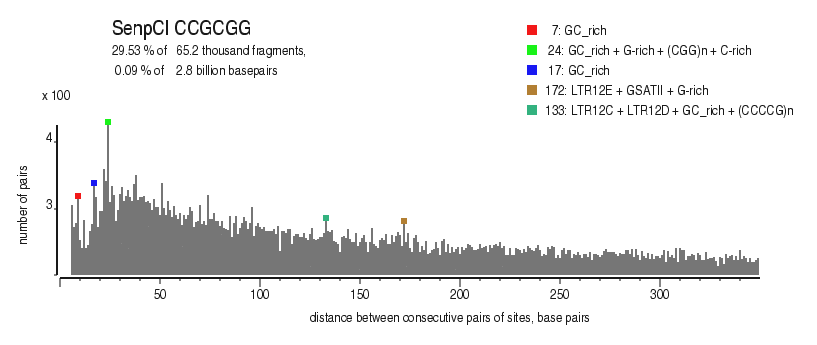
| Distribution of closely spaced sites |
|---|
 |
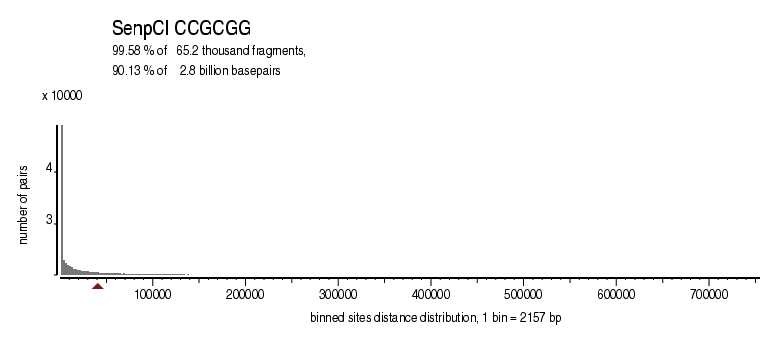
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 2312892 | chr2 | NT_005403.17 | 61299302-63612194 | 41.67 % in | 3443 repeats | 62.08 % in | 7 genes |
| 2 | 2066442 | chrX | NT_011669.17 | 3904892-5971334 | 72.93 % in | 3320 repeats | 28.97 % in | 8 genes |
| 3 | 1973303 | chr14 | NT_026437.12 | 65023241-66996544 | 45.18 % in | 3215 repeats | 0.35 % in | 4 genes |
| 4 | 1958699 | chr12 | NT_029419.12 | 45489151-47447850 | 53.67 % in | 2993 repeats | 12.03 % in | 4 genes |
| 5 | 1931649 | chr10 | NT_030059.13 | 57212777-59144426 | 47.16 % in | 3267 repeats | 32.46 % in | 4 genes |
| 6 | 1868985 | chr5 | NT_029289.11 | 3947948-5816933 | 53.42 % in | 3268 repeats | 20.07 % in | 8 genes |
| 7 | 1846168 | chrX | NT_011786.16 | 10114037-11960205 | 71.63 % in | 2626 repeats | 0.71 % in | 10 genes |
| 8 | 1793317 | chr12 | NT_029419.12 | 34809682-36602999 | 55.79 % in | 2723 repeats | 22.81 % in | 4 genes |
| 9 | 1713761 | chr2 | NT_022171.15 | 8027226-9740987 | 48.91 % in | 2558 repeats | 0.00 % in | 0 genes |
| 10 | 1713351 | chrX | NT_011651.17 | 16941077-18654428 | 73.53 % in | 2564 repeats | 0.00 % in | 0 genes |
| 11 | 1702946 | chr14 | NT_026437.12 | 29145534-30848480 | 55.92 % in | 2520 repeats | 0.00 % in | 0 genes |
| 12 | 1673158 | chrX | NT_011651.17 | 4363678-6036836 | 80.64 % in | 2443 repeats | 0.00 % in | 0 genes |
| 13 | 1661572 | chr10 | NT_030059.13 | 34467488-36129060 | 48.88 % in | 2694 repeats | 0.00 % in | 0 genes |
| 14 | 1645002 | chr18 | NT_010966.14 | 22005031-23650033 | 53.00 % in | 2636 repeats | 0.00 % in | 0 genes |
| 15 | 1618142 | chr1 | NT_032977.9 | 49998211-51616353 | 50.39 % in | 2535 repeats | 0.00 % in | 0 genes |
| 16 | 1597962 | chr4 | NT_006238.11 | 4496889-6094851 | 56.93 % in | 2549 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 2312892 | chr2 | NT_005403.17 | 61299302-63612194 | 3443 | 467 | AT_rich (402) | MIR (149) | MIRb (146) | |
| 2 | 2066442 | chrX | NT_011669.17 | 3904892-5971334 | 3320 | 394 | MIRb (187) | MIR (146) | AluSx (115) | |
| 3 | 1973303 | chr14 | NT_026437.12 | 65023241-66996544 | 3215 | 464 | AT_rich (307) | MIRb (132) | MIR (120) | |
| 4 | 1958699 | chr12 | NT_029419.12 | 45489151-47447850 | 2993 | 452 | AT_rich (307) | L2a (135) | MIRb (95) | |
| 5 | 1931649 | chr10 | NT_030059.13 | 57212777-59144426 | 3267 | 435 | MIRb (205) | MIR (183) | AT_rich (166) | |
| 6 | 1868985 | chr5 | NT_029289.11 | 3947948-5816933 | 3268 | 423 | MIRb (234) | L2a (207) | MIR (205) | |
| 7 | 1846168 | chrX | NT_011786.16 | 10114037-11960205 | 2626 | 411 | AT_rich (178) | L2a (63) | MIRb (62) | |
| 8 | 1793317 | chr12 | NT_029419.12 | 34809682-36602999 | 2723 | 458 | AT_rich (283) | L2a (124) | MIRb (93) | |
| 9 | 1713761 | chr2 | NT_022171.15 | 8027226-9740987 | 2558 | 380 | AT_rich (225) | MIRb (85) | AluSx (82) | |
| 10 | 1713351 | chrX | NT_011651.17 | 16941077-18654428 | 2564 | 412 | AT_rich (182) | (TA)n (64) | L1M1 (57) | |
| 11 | 1702946 | chr14 | NT_026437.12 | 29145534-30848480 | 2520 | 425 | AT_rich (247) | L2a (82) | AluSx (80) | |
| 12 | 1673158 | chrX | NT_011651.17 | 4363678-6036836 | 2443 | 381 | AT_rich (124) | L1PA15 (56) | AluSx (56) | |
| 13 | 1661572 | chr10 | NT_030059.13 | 34467488-36129060 | 2694 | 412 | AT_rich (203) | MIRb (174) | AluSx (105) | |
| 14 | 1645002 | chr18 | NT_010966.14 | 22005031-23650033 | 2636 | 425 | AT_rich (220) | MIR (92) | MIRb (90) | |
| 15 | 1618142 | chr1 | NT_032977.9 | 49998211-51616353 | 2535 | 435 | AT_rich (270) | L2a (103) | MIR (93) | |
| 16 | 1597962 | chr4 | NT_006238.11 | 4496889-6094851 | 2549 | 434 | AT_rich (276) | MIR (105) | L2a (95) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 2312892 | chr2 | NT_005403.17 | 61299302-63612194 | ACADL | long-chain_specific_acyl-CoA_dehydrogenase,_mitochondrial_precursor |
| MYL1 | myosin_light_chain_1/3,_skeletal_muscle_isoform_isoform_3f | |||||
| LANCL1 | lanC-like_protein_1 | |||||
| CPS1IT | CPS1_intronic_transcript_(non-protein_coding) | |||||
| LOC100420775 | ||||||
| RPS27P10 | ||||||
| MIR548F2 | microRNA_548f-2 | |||||
| 2 | 2066442 | chrX | NT_011669.17 | 3904892-5971334 | GPR165P | |
| LOC402408 | ||||||
| EDA2R | tumor_necrosis_factor_receptor_superfamily_member_27 | |||||
| LOC100129144 | zinc_finger_protein_681 | |||||
| LOC100288853 | ||||||
| AR | androgen_receptor_isoform_2 | |||||
| LOC100127902 | ||||||
| LOC100288917 | oligophrenin-1 | |||||
| 3 | 1973303 | chr14 | NT_026437.12 | 65023241-66996544 | LOC100421611 | |
| RNU3P3 | ||||||
| LOC100506731 | hypothetical_LOC100506731 | |||||
| FLRT2 | leucine-rich_repeat_transmembrane_protein_FLRT2_precursor | |||||
| 4 | 1958699 | chr12 | NT_029419.12 | 45489151-47447850 | TMTC2 | transmembrane_and_TPR_repeat-containing_protein_2 |
| RPL6P25 | ||||||
| LOC100128335 | ||||||
| SLC6A15 | orphan_sodium-_and_chloride-dependent_neurotransmitter_transporter_NTT73_isoform_2 | |||||
| 5 | 1931649 | chr10 | NT_030059.13 | 57212777-59144426 | LOC100505890 | hypothetical_LOC100505890 |
| YWHAZP5 | ||||||
| RPL23AP59 | ||||||
| SORCS1 | VPS10_domain-containing_receptor_SorCS1_isoform_a | |||||
| 6 | 1868985 | chr5 | NT_029289.11 | 3947948-5816933 | RPL7P21 | glucocorticoid_receptor_isoform_gamma |
| LOC100420513 | ||||||
| HMHB1 | minor_histocompatibility_protein_HB-1 | |||||
| YIPF5 | protein_YIPF5 | |||||
| KCTD16 | BTB/POZ_domain-containing_protein_KCTD16 | |||||
| LOC100128121 | ||||||
| LOC100132712 | ||||||
| ASS1P10 | ||||||
| 7 | 1846168 | chrX | NT_011786.16 | 10114037-11960205 | LOC100288421 | |
| CXorf64 | hypothetical_protein_LOC100130613 | |||||
| PNPLA10P | ||||||
| LOC100420320 | ||||||
| ACTRT1 | actin-related_protein_T1 | |||||
| LOC100129947 | ||||||
| LOC100419246 | ||||||
| LOC100128268 | ||||||
| RPL7AP72 | ||||||
| LOC392538 | ||||||
| 8 | 1793317 | chr12 | NT_029419.12 | 34809682-36602999 | LOC283392 | hypothetical_LOC283392,_transcript_variant_2 |
| LOC100422354 | thyrotropin-releasing_hormone-degrading_ectoenzyme | |||||
| LOC100507363 | hypothetical_LOC100507363 | |||||
| RPL31P48 | ||||||