| Enzyme: | ClaI | Longest uncut segments |
| Specificity: | ATCGAT | Repeats in uncut segments |
| Number of sites: | 85091 | Genes in uncut segments |
| Mean distance between sites: | 33626 base pairs | |
| Standard deviation: | 35270 base pairs | |
| Site density | 29.7 per megabase | Help |
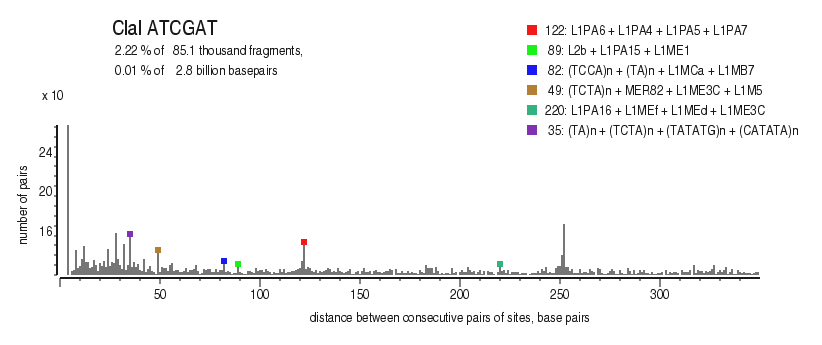
| Distribution of closely spaced sites |
|---|
 |
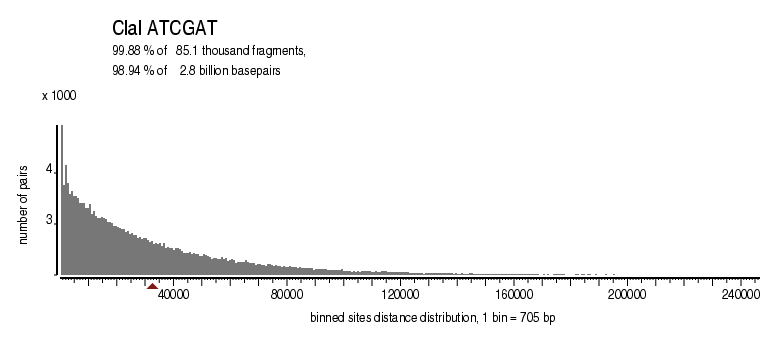
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 647243 | chr15 | NT_037852.6 | 1255774-1903017 | 13.86 % in | 381 repeats | 2.96 % in | 3 genes |
| 2 | 492771 | chr6 | NT_167244.1 | 1820515-2313286 | 14.91 % in | 299 repeats | 17.27 % in | 11 genes |
| 3 | 474874 | chr6 | NT_167244.1 | 2326429-2801303 | 7.85 % in | 149 repeats | 3.94 % in | 2 genes |
| 4 | 462928 | chr17 | NT_010783.15 | 5578220-6041148 | 46.91 % in | 1050 repeats | 78.40 % in | 18 genes |
| 5 | 412844 | chr2 | NT_005403.17 | 54359304-54772148 | 49.47 % in | 915 repeats | 55.29 % in | 6 genes |
| 6 | 412011 | chr15 | NT_010194.17 | 46235072-46647083 | 46.34 % in | 931 repeats | 61.66 % in | 12 genes |
| 7 | 399111 | chr19 | NT_011109.16 | 15465806-15864917 | 56.74 % in | 616 repeats | 52.26 % in | 8 genes |
| 8 | 391486 | chr9 | NT_008413.18 | 4701472-5092958 | 56.48 % in | 763 repeats | 56.02 % in | 6 genes |
| 9 | 381373 | chr20 | NT_011362.10 | 4899186-5280559 | 44.80 % in | 867 repeats | 0.00 % in | 0 genes |
| 10 | 356219 | chr12 | NT_009775.17 | 11259621-11615840 | 61.12 % in | 1113 repeats | 0.00 % in | 0 genes |
| 11 | 356063 | chr12 | NT_029419.12 | 33620090-33976153 | 58.55 % in | 566 repeats | 0.00 % in | 0 genes |
| 12 | 355768 | chr1 | NT_032977.9 | 22939245-23295013 | 67.01 % in | 1116 repeats | 0.00 % in | 0 genes |
| 13 | 354361 | chr6 | NT_167247.1 | 1401812-1756173 | 25.34 % in | 345 repeats | 0.00 % in | 0 genes |
| 14 | 348797 | chrY | NT_011875.12 | 8381724-8730521 | 81.63 % in | 106 repeats | 0.00 % in | 0 genes |
| 15 | 346749 | chr2 | NT_005403.17 | 29851154-30197903 | 43.27 % in | 464 repeats | 0.00 % in | 0 genes |
| 16 | 341449 | chr11 | NT_167190.1 | 4739868-5081317 | 55.21 % in | 672 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 647243 | chr15 | NT_037852.6 | 1255774-1903017 | 381 | 118 | AluSx (24) | MIRb (13) | AT_rich (13) | |
| 2 | 492771 | chr6 | NT_167244.1 | 1820515-2313286 | 299 | 87 | AluSx (41) | L2a (14) | AluSg (13) | |
| 3 | 474874 | chr6 | NT_167244.1 | 2326429-2801303 | 149 | 69 | AluSx (11) | AluY (10) | AluJo (8) | |
| 4 | 462928 | chr17 | NT_010783.15 | 5578220-6041148 | 1050 | 149 | AluSx (192) | AluJo (63) | AluY (57) | |
| 5 | 412844 | chr2 | NT_005403.17 | 54359304-54772148 | 915 | 187 | AluSx (125) | AluY (51) | AT_rich (50) | |
| 6 | 412011 | chr15 | NT_010194.17 | 46235072-46647083 | 931 | 178 | AluSx (111) | AluJo (62) | AluJb (62) | |
| 7 | 399111 | chr19 | NT_011109.16 | 15465806-15864917 | 616 | 91 | L1MB3 (32) | L1MB1 (30) | L1MCa (27) | |
| 8 | 391486 | chr9 | NT_008413.18 | 4701472-5092958 | 763 | 192 | AluSx (51) | AT_rich (46) | AluJo (39) | |
| 9 | 381373 | chr20 | NT_011362.10 | 4899186-5280559 | 867 | 141 | AluSx (83) | MIR (82) | MIRb (79) | |
| 10 | 356219 | chr12 | NT_009775.17 | 11259621-11615840 | 1113 | 150 | AluSx (160) | AluY (76) | AluJo (61) | |
| 11 | 356063 | chr12 | NT_029419.12 | 33620090-33976153 | 566 | 176 | AT_rich (33) | MIR (32) | L2a (29) | |
| 12 | 355768 | chr1 | NT_032977.9 | 22939245-23295013 | 1116 | 174 | AluSx (145) | AluJb (62) | AluJo (50) | |
| 13 | 354361 | chr6 | NT_167247.1 | 1401812-1756173 | 345 | 128 | L2a (18) | L2c (14) | MIRc (13) | |
| 14 | 348797 | chrY | NT_011875.12 | 8381724-8730521 | 106 | 48 | LTR12B (17) | L1PA16 (7) | AT_rich (7) | |
| 15 | 346749 | chr2 | NT_005403.17 | 29851154-30197903 | 464 | 141 | AT_rich (46) | MIRb (21) | L2c (21) | |
| 16 | 341449 | chr11 | NT_167190.1 | 4739868-5081317 | 672 | 185 | AluSx (47) | AT_rich (32) | MIRb (31) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 647243 | chr15 | NT_037852.6 | 1255774-1903017 | LOC283804 | |
| LOC727914 | ||||||
| LOC100418897 | ||||||
| 2 | 492771 | chr6 | NT_167244.1 | 1820515-2313286 | GNL1 | guanine_nucleotide-binding_protein-like_1 |
| DHX16 | putative_pre-mRNA-splicing_factor_ATP-dependent_RNA_helicase_DHX16_isoform_1 | |||||
| KIAA1949 | phostensin | |||||
| NRM | nurim | |||||
| RPL7P4 | ||||||
| MDC1 | mediator_of_DNA_damage_checkpoint_protein_1 | |||||
| LOC100294090 | hypothetical_LOC100294090,_transcript_variant_1 | |||||
| FLOT1 | flotillin-1 | |||||
| DDR1 | epithelial_discoidin_domain-containing_receptor_1_isoform_DDR1c | |||||
| MUC21 | mucin-21_precursor | |||||
| LOC100507702 | hypothetical_protein_LOC100507702 | |||||
| 3 | 474874 | chr6 | NT_167244.1 | 2326429-2801303 | HCG22 | HLA_complex_group_22 |
| MICB | MHC_class_I_polypeptide-related_sequence_B_precursor | |||||
| 4 | 462928 | chr17 | NT_010783.15 | 5578220-6041148 | RAB5C | ras-related_protein_Rab-5C_isoform_b |
| KCNH4 | potassium_voltage-gated_channel_subfamily_H_member_4 | |||||
| HCRT | orexin_precursor | |||||
| GHDC | GH3_domain-containing_protein_isoform_3 | |||||
| STAT5B | signal_transducer_and_activator_of_transcription_5B | |||||
| STAT5A | signal_transducer_and_activator_of_transcription_5A | |||||
| STAT3 | signal_transducer_and_activator_of_transcription_3_isoform_3 | |||||
| PTRF | polymerase_I_and_transcript_release_factor | |||||
| ATP6V0A1 | V-type_proton_ATPase_116_kDa_subunit_a_isoform_1_isoform_c | |||||
| PTP4A2P1 | ||||||
| NAGLU | alpha-N-acetylglucosaminidase_precursor | |||||
| HSD17B1P1 | ||||||
| HSD17B1 | estradiol_17-beta-dehydrogenase_1 | |||||
| COASY | bifunctional_coenzyme_A_synthase_isoform_b | |||||
| MLX | max-like_protein_X_isoform_gamma | |||||
| PSMC3IP | homologous-pairing_protein_2_homolog_isoform_2 | |||||
| FAM134C | hypothetical_protein_LOC162427 | |||||
| TUBG1 | tubulin_gamma-1_chain | |||||
| 5 | 412844 | chr2 | NT_005403.17 | 54359304-54772148 | CYP20A1 | cytochrome_P450_20A1 |
| MRPL50P2 | ||||||
| TRNAE40P | abl_interactor_2 | |||||
| RAPH1 | ras-associated_and_pleckstrin_homology_domains-containing_protein_1_isoform_3 | |||||
| LOC729532 | ||||||
| LOC100287498 | ||||||
| 6 | 412011 | chr15 | NT_010194.17 | 46235072-46647083 | RPL36AP45 | |
| C15orf39 | hypothetical_protein_LOC56905 | |||||
| LOC100128707 | ||||||
| LOC100506519 | putative_golgin_subfamily_A_member_6C | |||||
| LOC100506552 | golgin_A6_family,_member_D | |||||
| DNM1P34 | ||||||
| ANP32BP1 | ||||||
| COMMD4 | COMM_domain-containing_protein_4 | |||||
| MIR631 | microRNA:hsa-mir-631 | |||||
| MAN2C1 | alpha-mannosidase_2C1 | |||||
| RPL13P4 | paired_amphipathic_helix_protein_Sin3a | |||||
| LOC100506579 | hypothetical_protein_LOC100506579 | |||||
| 7 | 399111 | chr19 | NT_011109.16 | 15465806-15864917 | PSG3 | pregnancy-specific_beta-1-glycoprotein_3_precursor |
| LOC100505701 | hypothetical_LOC100505701 | |||||
| CEACAMP6 | ||||||
| PSG10 | pregnancy_specific_beta-1-glycoprotein_10 | |||||
| PSG1 | pregnancy-specific_beta-1-glycoprotein_1_isoform_1 | |||||
| PSG11 | pregnancy-specific_beta-1-glycoprotein_11_isoform_2 | |||||
| CEACAMP8 | ||||||
| PSG2 | pregnancy-specific_beta-1-glycoprotein_2_precursor | |||||
| 8 | 391486 | chr9 | NT_008413.18 | 4701472-5092958 | AK3 | GTP:AMP_phosphotransferase,_mitochondrial |
| LOC100420354 | ||||||
| RPS5P6 | ||||||
| MIR101-2 | microRNA:hsa-mir-101-2 | |||||
| LOC100128701 | ||||||
| IGHEP2 | tyrosine-protein_kinase_JAK2 | |||||