| Enzyme: | AfeI | Longest uncut segments |
| Specificity: | AGCGCT | Repeats in uncut segments |
| Number of sites: | 107947 | Genes in uncut segments |
| Mean distance between sites: | 26506 base pairs | |
| Standard deviation: | 34587 base pairs | |
| Site density | 37.7 per megabase | Help |
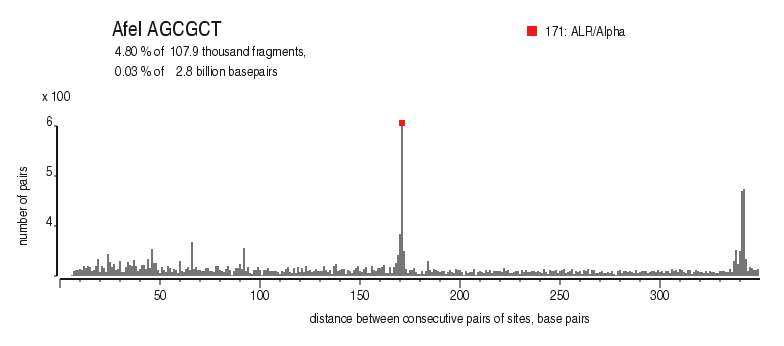
| Distribution of closely spaced sites |
|---|
 |
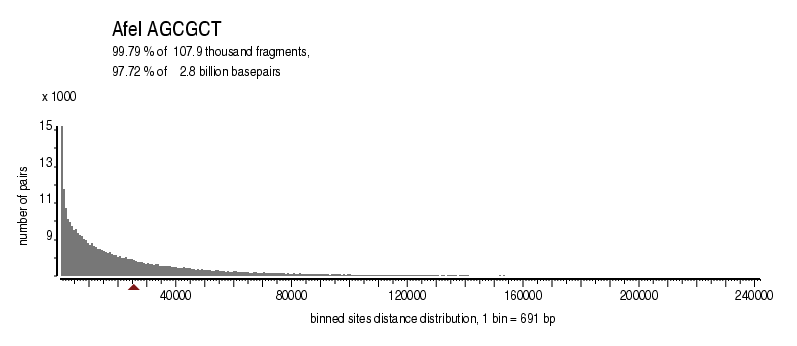
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 552209 | chr15 | NT_037852.6 | 1375681-1927890 | 5.79 % in | 126 repeats | 1.81 % in | 1 genes |
| 2 | 452884 | chr4 | NT_022778.16 | 4452573-4905457 | 57.69 % in | 709 repeats | 0.00 % in | 0 genes |
| 3 | 433662 | chr6 | NT_167244.1 | 2337194-2770856 | 3.87 % in | 72 repeats | 1.19 % in | 1 genes |
| 4 | 425597 | chr1 | NT_032977.9 | 73592189-74017786 | 45.47 % in | 657 repeats | 0.00 % in | 0 genes |
| 5 | 424975 | chr9 | NT_008413.18 | 25563501-25988476 | 54.97 % in | 655 repeats | 0.70 % in | 2 genes |
| 6 | 422461 | chr21 | NT_011512.11 | 8394785-8817246 | 46.75 % in | 675 repeats | 42.21 % in | 1 genes |
| 7 | 414615 | chr10 | NT_030059.13 | 35682443-36097058 | 65.00 % in | 676 repeats | 0.00 % in | 0 genes |
| 8 | 398471 | chr5 | NT_034772.6 | 33279226-33677697 | 54.97 % in | 532 repeats | 0.35 % in | 2 genes |
| 9 | 395823 | chr5 | NT_006576.16 | 18002271-18398094 | 49.61 % in | 646 repeats | 0.00 % in | 0 genes |
| 10 | 395689 | chrX | NT_011651.17 | 13168640-13564329 | 76.26 % in | 572 repeats | 0.00 % in | 0 genes |
| 11 | 394041 | chr3 | NT_022459.15 | 16734216-17128257 | 51.03 % in | 627 repeats | 0.00 % in | 0 genes |
| 12 | 393382 | chr14 | NT_026437.12 | 64667538-65060920 | 48.21 % in | 692 repeats | 0.00 % in | 0 genes |
| 13 | 392992 | chr17 | NT_010783.15 | 17291076-17684068 | 56.63 % in | 629 repeats | 0.00 % in | 0 genes |
| 14 | 377606 | chr11 | NT_009237.18 | 41445843-41823449 | 62.24 % in | 686 repeats | 0.00 % in | 0 genes |
| 15 | 375442 | chr7 | NT_007933.15 | 63282996-63658438 | 57.93 % in | 651 repeats | 0.00 % in | 0 genes |
| 16 | 375249 | chr3 | NT_005612.16 | 37717249-38092498 | 48.75 % in | 663 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 552209 | chr15 | NT_037852.6 | 1375681-1927890 | 126 | 62 | AT_rich (10) | Tigger2 (6) | L1MDa (6) | |
| 2 | 452884 | chr4 | NT_022778.16 | 4452573-4905457 | 709 | 206 | AT_rich (97) | (TA)n (27) | L2 (22) | |
| 3 | 433662 | chr6 | NT_167244.1 | 2337194-2770856 | 72 | 41 | AluSx (6) | AluJb (5) | L1ME4a (3) | |
| 4 | 425597 | chr1 | NT_032977.9 | 73592189-74017786 | 657 | 168 | AT_rich (74) | MIR (22) | L2a (20) | |
| 5 | 424975 | chr9 | NT_008413.18 | 25563501-25988476 | 655 | 203 | AT_rich (53) | L2a (31) | MIRb (28) | |
| 6 | 422461 | chr21 | NT_011512.11 | 8394785-8817246 | 675 | 202 | AT_rich (88) | AluSx (23) | L2a (21) | |
| 7 | 414615 | chr10 | NT_030059.13 | 35682443-36097058 | 676 | 222 | AT_rich (48) | MIRb (25) | MIR (24) | |
| 8 | 398471 | chr5 | NT_034772.6 | 33279226-33677697 | 532 | 184 | AT_rich (52) | MIRb (21) | L2a (19) | |
| 9 | 395823 | chr5 | NT_006576.16 | 18002271-18398094 | 646 | 207 | AT_rich (50) | L2a (30) | AluSx (22) | |
| 10 | 395689 | chrX | NT_011651.17 | 13168640-13564329 | 572 | 204 | AT_rich (53) | MIR (15) | L1PA16 (13) | |
| 11 | 394041 | chr3 | NT_022459.15 | 16734216-17128257 | 627 | 191 | AT_rich (94) | L2 (20) | (TA)n (19) | |
| 12 | 393382 | chr14 | NT_026437.12 | 64667538-65060920 | 692 | 218 | AT_rich (76) | MIRb (25) | MIR (24) | |
| 13 | 392992 | chr17 | NT_010783.15 | 17291076-17684068 | 629 | 193 | AT_rich (46) | L2a (33) | MIRb (27) | |
| 14 | 377606 | chr11 | NT_009237.18 | 41445843-41823449 | 686 | 211 | AT_rich (37) | MIRb (29) | MIR (25) | |
| 15 | 375442 | chr7 | NT_007933.15 | 63282996-63658438 | 651 | 207 | AT_rich (79) | L2a (23) | (TA)n (22) | |
| 16 | 375249 | chr3 | NT_005612.16 | 37717249-38092498 | 663 | 204 | MIRb (58) | MIR (32) | L2c (32) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 552209 | chr15 | NT_037852.6 | 1375681-1927890 | LOC100418897 | |
| 3 | 433662 | chr6 | NT_167244.1 | 2337194-2770856 | HCG22 | HLA_complex_group_22 |
| 5 | 424975 | chr9 | NT_008413.18 | 25563501-25988476 | TUSC1 | tumor_suppressor_candidate_gene_1_protein |
| LOC100421478 | ||||||
| 6 | 422461 | chr21 | NT_011512.11 | 8394785-8817246 | NCAM2 | neural_cell_adhesion_molecule_2_precursor |
| 8 | 398471 | chr5 | NT_034772.6 | 33279226-33677697 | LOC100419321 | |
| RPSAP37 | ||||||