| Enzyme: | MluI | Longest uncut segments |
| Specificity: | ACGCGT | Repeats in uncut segments |
| Number of sites: | 20881 | Genes in uncut segments |
| Mean distance between sites: | 137031 base pairs | |
| Standard deviation: | 192274 base pairs | |
| Site density | 7.3 per megabase | Help |
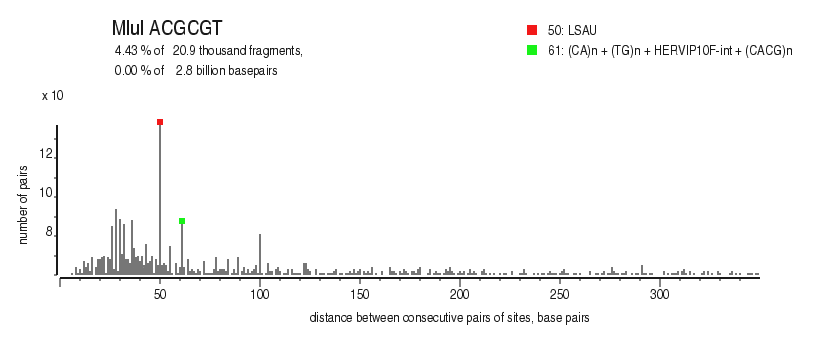
| Distribution of closely spaced sites |
|---|
 |
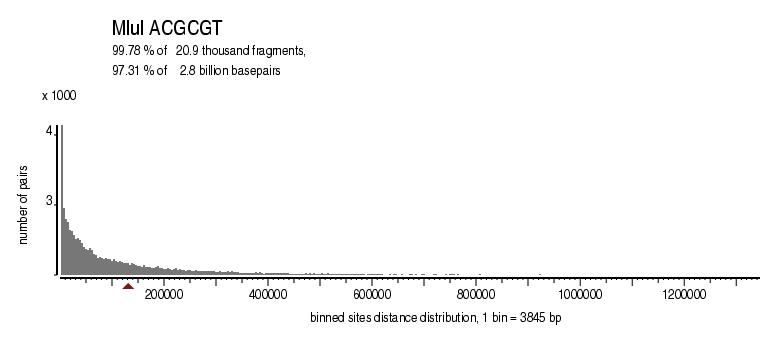
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 2276590 | chr2 | NT_005403.17 | 34535957-36812547 | 54.92 % in | 3460 repeats | 15.14 % in | 4 genes |
| 2 | 2092979 | chr4 | NT_016354.19 | 49227869-51320848 | 45.84 % in | 3236 repeats | 19.16 % in | 8 genes |
| 3 | 2068381 | chr5 | NT_034772.6 | 10163284-12231665 | 45.12 % in | 3180 repeats | 20.07 % in | 7 genes |
| 4 | 1979523 | chr3 | NT_005612.16 | 70650585-72630108 | 51.80 % in | 3182 repeats | 8.83 % in | 4 genes |
| 5 | 1971179 | chr3 | NT_022517.18 | 33823205-35794384 | 52.04 % in | 3083 repeats | 10.18 % in | 5 genes |
| 6 | 1957495 | chr2 | NT_005403.17 | 30097092-32054587 | 51.63 % in | 2987 repeats | 34.44 % in | 8 genes |
| 7 | 1935455 | chr3 | NT_005612.16 | 83803443-85738898 | 51.32 % in | 3346 repeats | 38.86 % in | 9 genes |
| 8 | 1886794 | chr4 | NT_016354.19 | 3779525-5666319 | 48.76 % in | 3025 repeats | 60.59 % in | 16 genes |
| 9 | 1866101 | chr11 | NT_009237.18 | 38389262-40255363 | 53.34 % in | 3055 repeats | 0.00 % in | 0 genes |
| 10 | 1828954 | chrX | NT_011651.17 | 17273966-19102920 | 70.48 % in | 2760 repeats | 0.00 % in | 0 genes |
| 11 | 1820311 | chr10 | NT_030059.13 | 59765062-61585373 | 53.34 % in | 2955 repeats | 0.00 % in | 0 genes |
| 12 | 1809187 | chr7 | NT_007933.15 | 22848515-24657702 | 51.17 % in | 2723 repeats | 0.00 % in | 0 genes |
| 13 | 1780872 | chr10 | NT_030059.13 | 4877668-6658540 | 51.33 % in | 2751 repeats | 0.00 % in | 0 genes |
| 14 | 1736127 | chr14 | NT_026437.12 | 61664025-63400152 | 50.60 % in | 2920 repeats | 0.00 % in | 0 genes |
| 15 | 1703976 | chr6 | NT_025741.15 | 6117116-7821092 | 50.09 % in | 2658 repeats | 0.00 % in | 0 genes |
| 16 | 1697641 | chr18 | NT_010966.14 | 8450302-10147943 | 51.77 % in | 2741 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 2276590 | chr2 | NT_005403.17 | 34535957-36812547 | 3460 | 512 | AT_rich (440) | L2a (95) | AluSx (83) | |
| 2 | 2092979 | chr4 | NT_016354.19 | 49227869-51320848 | 3236 | 482 | AT_rich (379) | MIR (141) | L2a (130) | |
| 3 | 2068381 | chr5 | NT_034772.6 | 10163284-12231665 | 3180 | 454 | AT_rich (340) | MIRb (122) | L2a (122) | |
| 4 | 1979523 | chr3 | NT_005612.16 | 70650585-72630108 | 3182 | 448 | AT_rich (413) | L2a (99) | (TA)n (84) | |
| 5 | 1971179 | chr3 | NT_022517.18 | 33823205-35794384 | 3083 | 437 | AT_rich (197) | MIRb (165) | MIR (109) | |
| 6 | 1957495 | chr2 | NT_005403.17 | 30097092-32054587 | 2987 | 434 | AT_rich (232) | L2a (129) | MIR (122) | |
| 7 | 1935455 | chr3 | NT_005612.16 | 83803443-85738898 | 3346 | 455 | AT_rich (222) | MIRb (197) | AluSx (142) | |
| 8 | 1886794 | chr4 | NT_016354.19 | 3779525-5666319 | 3025 | 452 | AT_rich (240) | L2a (151) | MIRb (130) | |
| 9 | 1866101 | chr11 | NT_009237.18 | 38389262-40255363 | 3055 | 452 | AT_rich (285) | MIRb (149) | AluY (137) | |
| 10 | 1828954 | chrX | NT_011651.17 | 17273966-19102920 | 2760 | 438 | AT_rich (181) | AluSx (67) | MIRb (65) | |
| 11 | 1820311 | chr10 | NT_030059.13 | 59765062-61585373 | 2955 | 421 | MIRb (194) | AT_rich (167) | MIR (130) | |
| 12 | 1809187 | chr7 | NT_007933.15 | 22848515-24657702 | 2723 | 421 | AT_rich (279) | L2a (98) | MIR (86) | |
| 13 | 1780872 | chr10 | NT_030059.13 | 4877668-6658540 | 2751 | 446 | AT_rich (250) | MIR (120) | MIRb (119) | |
| 14 | 1736127 | chr14 | NT_026437.12 | 61664025-63400152 | 2920 | 440 | AT_rich (152) | AluSx (144) | MIRb (136) | |
| 15 | 1703976 | chr6 | NT_025741.15 | 6117116-7821092 | 2658 | 422 | AT_rich (378) | L2a (84) | (TA)n (75) | |
| 16 | 1697641 | chr18 | NT_010966.14 | 8450302-10147943 | 2741 | 450 | AT_rich (308) | L2a (90) | MIR (84) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 2276590 | chr2 | NT_005403.17 | 34535957-36812547 | LOC644877 | |
| RPL23AP33 | zinc_finger_protein_804A | |||||
| LOC100506923 | hypothetical_LOC100506923 | |||||
| ELF2P4 | ||||||
| 2 | 2092979 | chr4 | NT_016354.19 | 49227869-51320848 | LOC285419 | hypothetical_LOC285419,_transcript_variant_2 |
| LOC645773 | ||||||
| LOC391696 | ||||||
| ANKRD50 | ankyrin_repeat_domain-containing_protein_50_isoform_2 | |||||
| FAT4 | protocadherin_Fat_4_precursor | |||||
| MIR2054 | microRNA_2054 | |||||
| LOC100289295 | ||||||
| LOC645841 | ||||||
| 3 | 2068381 | chr5 | NT_034772.6 | 10163284-12231665 | PAM | peptidyl-glycine_alpha-amidating_monooxygenase_isoform_e_preproprotein |
| LOC134505 | ||||||
| GIN1 | gypsy_retrotransposon_integrase-like_protein_1 | |||||
| PPIP5K2 | inositol_hexakisphosphate_and_diphosphoinositol-pentakisphosphate_kinase_2 | |||||
| C5orf30 | hypothetical_protein_LOC90355 | |||||
| LOC100129962 | ||||||
| NUDT12 | peroxisomal_NADH_pyrophosphatase_NUDT12 | |||||
| 4 | 1979523 | chr3 | NT_005612.16 | 70650585-72630108 | SI | sucrase-isomaltase,_intestinal |
| SLITRK3 | SLIT_and_NTRK-like_protein_3_precursor | |||||
| BCHE | cholinesterase_precursor | |||||
| LOC100420620 | ||||||
| 5 | 1971179 | chr3 | NT_022517.18 | 33823205-35794384 | PDCD6IP | programmed_cell_death_6_interacting_protein,_transcript_variant_3 |
| FECHP1 | ||||||
| KRT8P18 | ||||||
| LOC100130503 | hypothetical_protein_LOC100130503 | |||||
| MIR128-2 | microRNA:hsa-mir-128-2 | |||||
| 6 | 1957495 | chr2 | NT_005403.17 | 30097092-32054587 | RPS6P2 | coiled-coil_domain-containing_protein_141 |
| SESTD1 | SEC14_domain_and_spectrin_repeat-containing_protein_1 | |||||
| LOC644776 | ||||||
| MIR1258 | microRNA:hsa-mir-1258 | |||||
| CWC22 | pre-mRNA-splicing_factor_CWC22_homolog | |||||
| FTH1P20 | ||||||
| RPL27AP3 | ||||||
| UBE2E3 | ubiquitin-conjugating_enzyme_E2_E3 | |||||
| 7 | 1935455 | chr3 | NT_005612.16 | 83803443-85738898 | FGFR3P4 | hypothetical_LOC100505566,_transcript_variant_2 |
| KCNMB2 | calcium-activated_potassium_channel_subunit_beta-2 | |||||
| ZMAT3 | zinc_finger_matrin-type_protein_3_isoform_2 | |||||
| PIK3CA | phosphatidylinositol-4,5-bisphosphate_3-kinase_catalytic_subunit_alpha_isoform | |||||
| KCNMB3 | calcium-activated_potassium_channel_subunit_beta-3_isoform_c | |||||
| ZNF639 | zinc_finger_protein_639 | |||||
| MFN1 | mitofusin-1 | |||||
| GNB4 | guanine_nucleotide-binding_protein_subunit_beta-4 | |||||
| LOC442098 | ||||||
| 8 | 1886794 | chr4 | NT_016354.19 | 3779525-5666319 | LOC100462957 | extracellular_matrix_protein_FRAS1_isoform_2_precursor |
| ANXA3 | annexin_A3 | |||||
| LOC100505702 | hypothetical_LOC100505702,_transcript_variant_2 | |||||
| RPS21P3 | ||||||
| BMP2K | BMP-2-inducible_protein_kinase_isoform_b | |||||
| PAQR3 | progestin_and_adipoQ_receptor_family_member_3 | |||||
| LOC100505875 | hypothetical_LOC100505875,_transcript_variant_2 | |||||
| NAA11 | N-alpha-acetyltransferase_11,_NatA_catalytic_subunit | |||||
| GK2 | glycerol_kinase_2 | |||||
| LOC100506035 | hypothetical_LOC100506035,_transcript_variant_1 | |||||
| OR7E94P | ||||||
| GDEP | gene_differentially_expressed_in_prostate | |||||
| ANTXR2 | anthrax_toxin_receptor_2_isoform_2 | |||||
| LOC643513 | ||||||
| RPSAP39 | ||||||
| PRDM8 | PR_domain_zinc_finger_protein_8 | |||||