| Enzyme: | AgeI | Longest uncut segments |
| Specificity: | ACCGGT | Repeats in uncut segments |
| Number of sites: | 52697 | Genes in uncut segments |
| Mean distance between sites: | 54298 base pairs | |
| Standard deviation: | 59099 base pairs | |
| Site density | 18.4 per megabase | Help |
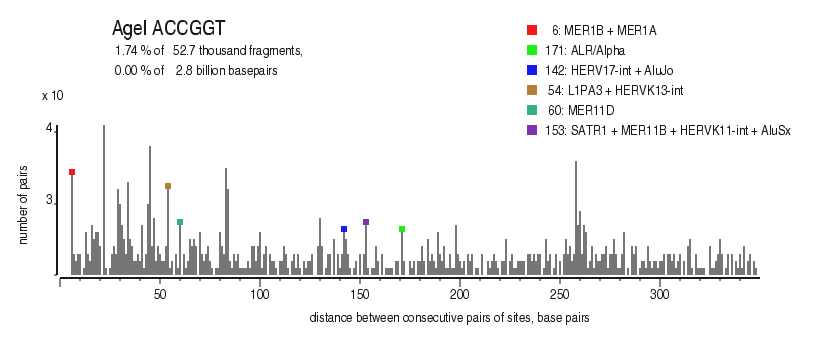
| Distribution of closely spaced sites |
|---|
 |
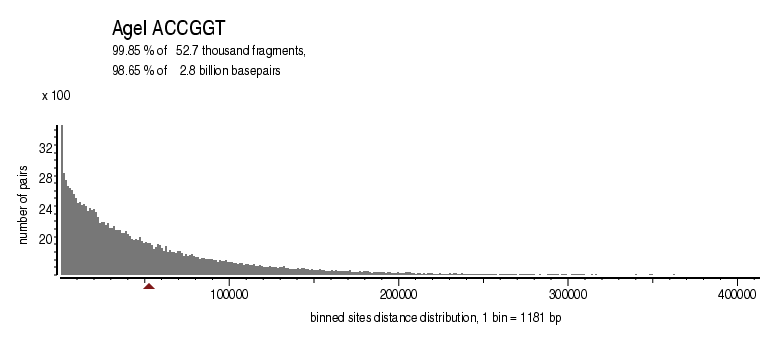
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 812581 | chr6 | NT_167244.1 | 1999507-2812088 | 11.51 % in | 348 repeats | 6.05 % in | 10 genes |
| 2 | 634570 | chr4 | NT_016354.19 | 48901497-49536067 | 48.36 % in | 1055 repeats | 43.75 % in | 2 genes |
| 3 | 631328 | chr6 | NT_007299.13 | 23721372-24352700 | 51.30 % in | 917 repeats | 13.11 % in | 7 genes |
| 4 | 605970 | chr13 | NT_024524.14 | 60925208-61531178 | 54.03 % in | 939 repeats | 21.07 % in | 3 genes |
| 5 | 593915 | chr13 | NT_024524.14 | 45594364-46188279 | 50.27 % in | 935 repeats | 0.00 % in | 0 genes |
| 6 | 572296 | chrX | NT_011786.16 | 26235183-26807479 | 62.77 % in | 963 repeats | 1.57 % in | 2 genes |
| 7 | 571998 | chr17 | NT_010783.15 | 26104542-26676540 | 57.52 % in | 1155 repeats | 65.01 % in | 5 genes |
| 8 | 566673 | chr6 | NT_007299.13 | 10445884-11012557 | 51.21 % in | 835 repeats | 52.25 % in | 1 genes |
| 9 | 559045 | chr11 | NT_167190.1 | 39838078-40397123 | 42.31 % in | 915 repeats | 0.00 % in | 0 genes |
| 10 | 548648 | chr2 | NT_022184.15 | 57213110-57761758 | 56.63 % in | 912 repeats | 0.00 % in | 0 genes |
| 11 | 546046 | chr5 | NT_034772.6 | 14285742-14831788 | 45.16 % in | 752 repeats | 0.00 % in | 0 genes |
| 12 | 545758 | chr7 | NT_033968.6 | 3272715-3818473 | 56.12 % in | 846 repeats | 0.00 % in | 0 genes |
| 13 | 541008 | chr13 | NT_024524.14 | 34794581-35335589 | 43.30 % in | 841 repeats | 0.00 % in | 0 genes |
| 14 | 540141 | chr8 | NT_008046.16 | 27139277-27679418 | 41.81 % in | 864 repeats | 0.00 % in | 0 genes |
| 15 | 529813 | chr8 | NT_008046.16 | 43488698-44018511 | 55.02 % in | 1103 repeats | 0.00 % in | 0 genes |
| 16 | 528021 | chr15 | NT_037852.6 | 1396122-1924143 | 2.84 % in | 71 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 812581 | chr6 | NT_167244.1 | 1999507-2812088 | 348 | 120 | AluSx (36) | AluY (14) | AluSp (13) | |
| 2 | 634570 | chr4 | NT_016354.19 | 48901497-49536067 | 1055 | 267 | AT_rich (92) | AluSx (46) | MIRb (42) | |
| 3 | 631328 | chr6 | NT_007299.13 | 23721372-24352700 | 917 | 254 | AT_rich (43) | MIR (36) | L2c (35) | |
| 4 | 605970 | chr13 | NT_024524.14 | 60925208-61531178 | 939 | 252 | AT_rich (61) | AluSx (40) | MIR (37) | |
| 5 | 593915 | chr13 | NT_024524.14 | 45594364-46188279 | 935 | 256 | AT_rich (124) | (TA)n (30) | MIR (28) | |
| 6 | 572296 | chrX | NT_011786.16 | 26235183-26807479 | 963 | 253 | AT_rich (54) | MIRb (46) | L2a (38) | |
| 7 | 571998 | chr17 | NT_010783.15 | 26104542-26676540 | 1155 | 242 | AluSx (114) | AT_rich (48) | AluJo (46) | |
| 8 | 566673 | chr6 | NT_007299.13 | 10445884-11012557 | 835 | 224 | AT_rich (78) | MIRb (42) | L2a (33) | |
| 9 | 559045 | chr11 | NT_167190.1 | 39838078-40397123 | 915 | 225 | MIRb (63) | L2a (49) | MIR (46) | |
| 10 | 548648 | chr2 | NT_022184.15 | 57213110-57761758 | 912 | 248 | AT_rich (112) | (TA)n (22) | L2a (22) | |
| 11 | 546046 | chr5 | NT_034772.6 | 14285742-14831788 | 752 | 219 | AT_rich (102) | L2a (36) | MIR (31) | |
| 12 | 545758 | chr7 | NT_033968.6 | 3272715-3818473 | 846 | 247 | AT_rich (81) | AluSx (28) | L1M5 (21) | |
| 13 | 541008 | chr13 | NT_024524.14 | 34794581-35335589 | 841 | 244 | AT_rich (56) | MIRb (48) | MIR (47) | |
| 14 | 540141 | chr8 | NT_008046.16 | 27139277-27679418 | 864 | 226 | AT_rich (112) | L2a (47) | L2c (40) | |
| 15 | 529813 | chr8 | NT_008046.16 | 43488698-44018511 | 1103 | 240 | MIRb (117) | MIR (57) | L2b (56) | |
| 16 | 528021 | chr15 | NT_037852.6 | 1396122-1924143 | 71 | 42 | AT_rich (8) | L2a (5) | L1MC4 (4) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 812581 | chr6 | NT_167244.1 | 1999507-2812088 | LOC100294090 | hypothetical_LOC100294090,_transcript_variant_1 |
| FLOT1 | flotillin-1 | |||||
| DDR1 | epithelial_discoidin_domain-containing_receptor_1_isoform_DDR1c | |||||
| MUC21 | mucin-21_precursor | |||||
| LOC100507702 | hypothetical_protein_LOC100507702 | |||||
| HCG22 | HLA_complex_group_22 | |||||
| MICB | MHC_class_I_polypeptide-related_sequence_B_precursor | |||||
| PPIAP9 | ||||||
| RPL15P4 | ||||||
| MCCD1 | mitochondrial_coiled-coil_domain_protein_1_precursor | |||||
| 2 | 634570 | chr4 | NT_016354.19 | 48901497-49536067 | - | tRNA-Cys |
| LOC285419 | hypothetical_LOC285419,_transcript_variant_2 | |||||
| 3 | 631328 | chr6 | NT_007299.13 | 23721372-24352700 | LOC100421583 | |
| RPL31P32 | ||||||
| KRT18P30 | ||||||
| LOC100421093 | ||||||
| LOC643870 | ||||||
| NT5E | 5'-nucleotidase_precursor | |||||
| SNX14 | sorting_nexin-14_isoform_b | |||||
| 4 | 605970 | chr13 | NT_024524.14 | 60925208-61531178 | RBM26 | RNA-binding_protein_26 |
| LOC100505538 | hypothetical_LOC100505538 | |||||
| NDFIP2 | NEDD4_family-interacting_protein_2_isoform_2 | |||||
| 6 | 572296 | chrX | NT_011786.16 | 26235183-26807479 | SPANXN4 | sperm_protein_associated_with_the_nucleus_on_the_X_chromosome_N4 |
| LOC100421180 | ||||||
| 7 | 571998 | chr17 | NT_010783.15 | 26104542-26676540 | MARCH10 | probable_E3_ubiquitin-protein_ligase_MARCH10 |
| LOC100128712 | ||||||
| MIR633 | microRNA:hsa-mir-633 | |||||
| LOC729667 | ||||||
| LOC100420064 | protein_TANC2 | |||||
| 8 | 566673 | chr6 | NT_007299.13 | 10445884-11012557 | RIMS1 | regulating_synaptic_membrane_exocytosis_protein_1_isoform_6 |