| Enzyme: | AclI | Longest uncut segments |
| Specificity: | AACGTT | Repeats in uncut segments |
| Number of sites: | 162140 | Genes in uncut segments |
| Mean distance between sites: | 17647 base pairs | |
| Standard deviation: | 18473 base pairs | |
| Site density | 56.7 per megabase | Help |
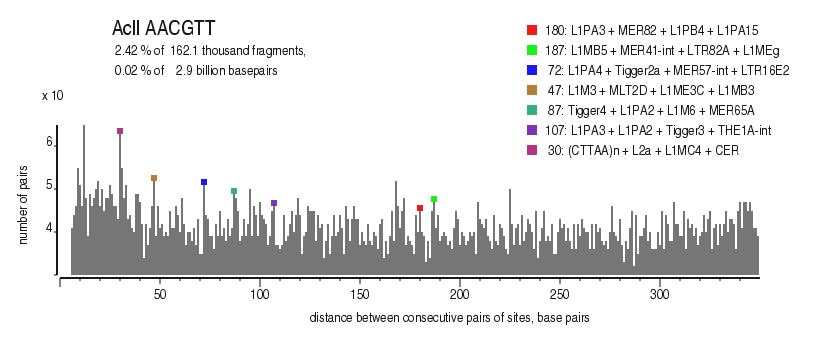
| Distribution of closely spaced sites |
|---|
 |
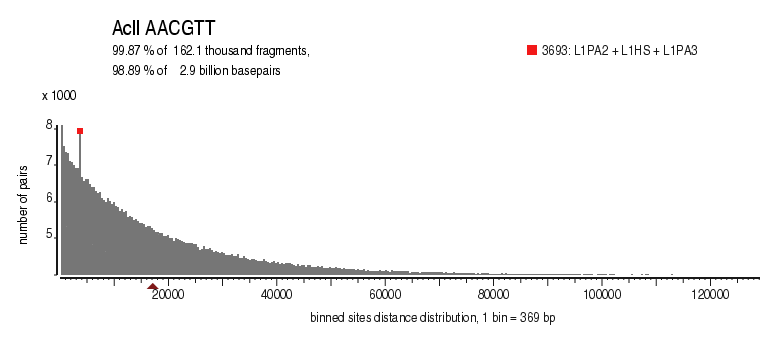
| Distribution of sites within 7 STD distance |
 |
| Help | ||||||||
|---|---|---|---|---|---|---|---|---|
| Longest uncut segments | ||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeat content | Gene content | ||
| 1 | 517786 | chr15 | NT_037852.6 | 1376835-1894621 | 3.92 % in | 68 repeats | 0.30 % in | 1 genes |
| 2 | 440642 | chr6 | NT_167246.1 | 3050489-3491131 | 14.84 % in | 302 repeats | 22.35 % in | 11 genes |
| 3 | 432033 | chr6 | NT_167244.1 | 2353750-2785783 | 3.62 % in | 63 repeats | 1.21 % in | 1 genes |
| 4 | 314469 | chr6 | NT_167244.1 | 3148579-3463048 | 12.07 % in | 193 repeats | 23.65 % in | 9 genes |
| 5 | 272590 | chr6 | NT_167247.1 | 4383440-4656030 | 9.12 % in | 101 repeats | 89.60 % in | 10 genes |
| 6 | 266078 | chr6 | NT_167244.1 | 1994698-2260776 | 4.29 % in | 55 repeats | 5.65 % in | 4 genes |
| 7 | 262379 | chr5 | NT_034772.6 | 16032453-16294832 | 65.29 % in | 465 repeats | 0.20 % in | 1 genes |
| 8 | 262206 | chr6 | NT_167247.1 | 1065601-1327807 | 33.19 % in | 260 repeats | 21.39 % in | 23 genes |
| 9 | 253407 | chr6 | NT_167247.1 | 1477096-1730503 | 16.61 % in | 176 repeats | 0.00 % in | 0 genes |
| 10 | 246332 | chr1 | NT_004487.19 | 6366873-6613205 | 38.26 % in | 495 repeats | 0.00 % in | 0 genes |
| 11 | 241657 | chr6 | NT_167244.1 | 2803351-3045008 | 12.75 % in | 156 repeats | 0.00 % in | 0 genes |
| 12 | 232123 | chr11 | NT_167190.1 | 17556468-17788591 | 34.05 % in | 424 repeats | 0.00 % in | 0 genes |
| 13 | 221751 | chr10 | NT_030059.13 | 55047421-55269172 | 45.37 % in | 466 repeats | 0.00 % in | 0 genes |
| 14 | 219965 | chr6 | NT_167244.1 | 4387352-4607317 | 4.95 % in | 21 repeats | 0.00 % in | 0 genes |
| 15 | 209068 | chr11 | NT_009237.18 | 15086907-15295975 | 40.85 % in | 393 repeats | 0.00 % in | 0 genes |
| 16 | 208598 | chr17 | NT_010783.15 | 38851377-39059975 | 37.28 % in | 401 repeats | 0.00 % in | 0 genes |
| Help | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeats in longest uncut segments | ||||||||||
| # | Length | Chr | Scaffold | Coordinates | Repeats | |||||
| Total | Distinct | Most | Second | Third | ||||||
| 1 | 517786 | chr15 | NT_037852.6 | 1376835-1894621 | 68 | 39 | Tigger2 (6) | L1MDa (6) | AluSx (4) | |
| 2 | 440642 | chr6 | NT_167246.1 | 3050489-3491131 | 302 | 84 | AluSx (48) | AluY (19) | AluSq (15) | |
| 3 | 432033 | chr6 | NT_167244.1 | 2353750-2785783 | 63 | 36 | AluY (6) | L4 (3) | L1ME4a (3) | |
| 4 | 314469 | chr6 | NT_167244.1 | 3148579-3463048 | 193 | 55 | AluSx (25) | MIR (11) | AluSq (11) | |
| 5 | 272590 | chr6 | NT_167247.1 | 4383440-4656030 | 101 | 56 | MIRb (5) | MIR (5) | L1PB1 (5) | |
| 6 | 266078 | chr6 | NT_167244.1 | 1994698-2260776 | 55 | 30 | AluSx (7) | MIRb (3) | MIR (3) | |
| 7 | 262379 | chr5 | NT_034772.6 | 16032453-16294832 | 465 | 181 | AT_rich (22) | AluJo (15) | L2a (13) | |
| 8 | 262206 | chr6 | NT_167247.1 | 1065601-1327807 | 260 | 106 | ERV3-16A3_I-int (12) | Charlie9 (12) | MLT1F1 (9) | |
| 9 | 253407 | chr6 | NT_167247.1 | 1477096-1730503 | 176 | 77 | L2a (12) | MIR3 (10) | L2c (9) | |
| 10 | 246332 | chr1 | NT_004487.19 | 6366873-6613205 | 495 | 102 | AluSx (68) | L2c (29) | AluJo (29) | |
| 11 | 241657 | chr6 | NT_167244.1 | 2803351-3045008 | 156 | 48 | AluSx (22) | AluJo (15) | AluJb (10) | |
| 12 | 232123 | chr11 | NT_167190.1 | 17556468-17788591 | 424 | 91 | MIRb (44) | L2b (39) | MIR (35) | |
| 13 | 221751 | chr10 | NT_030059.13 | 55047421-55269172 | 466 | 102 | AluSx (68) | AluJb (33) | AluY (30) | |
| 14 | 219965 | chr6 | NT_167244.1 | 4387352-4607317 | 21 | 14 | HERVH-int (4) | AluSx (3) | MER57-int (2) | |
| 15 | 209068 | chr11 | NT_009237.18 | 15086907-15295975 | 393 | 115 | MIRb (60) | MIR (36) | L2c (26) | |
| 16 | 208598 | chr17 | NT_010783.15 | 38851377-39059975 | 401 | 83 | AluSx (61) | AluY (28) | AluJo (27) | |
| Help | ||||||
|---|---|---|---|---|---|---|
| Genes in longest uncut segments | ||||||
| Sgmnt | Length (bp) | Chr | Scaffold | Coordinates | Gene symbol | Gene function |
| 1 | 517786 | chr15 | NT_037852.6 | 1376835-1894621 | LOC100418897 | |
| 2 | 440642 | chr6 | NT_167246.1 | 3050489-3491131 | MSH5 | mutS_protein_homolog_5_isoform_c |
| NEU1 | sialidase-1_precursor | |||||
| C2 | complement_C2_isoform_3 | |||||
| CFB | complement_factor_B_preproprotein_preproprotein | |||||
| TNXB | tenascin-X_isoform_1_precursor | |||||
| PRRT1 | proline-rich_transmembrane_protein_1 | |||||
| LOC100294186 | hypothetical_LOC100294186,_transcript_variant_2 | |||||
| PPT2 | lysosomal_thioesterase_PPT2_isoform_a_precursor | |||||
| EGFL8 | epidermal_growth_factor-like_protein_8_precursor | |||||
| AGPAT1 | 1-acyl-sn-glycerol-3-phosphate_acyltransferase_alpha | |||||
| RNF5 | E3_ubiquitin-protein_ligase_RNF5 | |||||
| 3 | 432033 | chr6 | NT_167244.1 | 2353750-2785783 | MICB | MHC_class_I_polypeptide-related_sequence_B_precursor |
| 4 | 314469 | chr6 | NT_167244.1 | 3148579-3463048 | SLC44A4 | choline_transporter-like_protein_4_isoform_3 |
| EHMT2 | histone-lysine_N-methyltransferase,_H3_lysine-9_specific_3_isoform_b | |||||
| TNXB | tenascin-X_isoform_1_precursor | |||||
| PRRT1 | proline-rich_transmembrane_protein_1 | |||||
| LOC100294105 | hypothetical_LOC100294105,_transcript_variant_2 | |||||
| PPT2 | lysosomal_thioesterase_PPT2_isoform_a_precursor | |||||
| EGFL8 | epidermal_growth_factor-like_protein_8_precursor | |||||
| AGPAT1 | 1-acyl-sn-glycerol-3-phosphate_acyltransferase_alpha | |||||
| RNF5 | E3_ubiquitin-protein_ligase_RNF5 | |||||
| 5 | 272590 | chr6 | NT_167247.1 | 4383440-4656030 | RPL32P1 | HLA_class_II_histocompatibility_antigen,_DP_beta_1_chain_precursor |
| HLA-DPA2 | ||||||
| COL11A2P | ||||||
| LOC100507722 | hypothetical_protein_LOC100507722 | |||||
| COL11A2 | collagen_alpha-2(XI)_chain_isoform_4_precursor | |||||
| RNY4P10 | retinoic_acid_receptor_RXR-beta | |||||
| SLC39A7 | zinc_transporter_SLC39A7 | |||||
| HSD17B8 | estradiol_17-beta-dehydrogenase_8 | |||||
| MIR219-1 | microRNA:hsa-mir-219-1 | |||||
| RING1 | E3_ubiquitin-protein_ligase_RING1 | |||||
| 6 | 266078 | chr6 | NT_167244.1 | 1994698-2260776 | MDC1 | mediator_of_DNA_damage_checkpoint_protein_1 |
| LOC100294090 | hypothetical_LOC100294090,_transcript_variant_1 | |||||
| FLOT1 | flotillin-1 | |||||
| DDR1 | epithelial_discoidin_domain-containing_receptor_1_isoform_DDR1c | |||||
| 7 | 262379 | chr5 | NT_034772.6 | 16032453-16294832 | RPS20P3 | |
| 8 | 262206 | chr6 | NT_167247.1 | 1065601-1327807 | HCG4P9 | |
| HLA-P | ||||||
| RPL7AP7 | ||||||
| HCG2P8 | ||||||
| MICG | ||||||
| HCP5P12 | ||||||
| HCG4P8 | ||||||
| HLA-G | HLA_class_I_histocompatibility_antigen,_alpha_chain_G_precursor | |||||
| LOC100133214 | hypothetical_protein_LOC100133214 | |||||
| HCGVIII-2 | ||||||
| MICF | ||||||
| 3.8-1.4 | ||||||
| HCG4P7 | ||||||
| P5-09 | major_histocompatibility_complex,_class_I,_H_(pseudogene) | |||||
| P5-07 | ||||||
| HLA-T | ||||||
| HCG2P7 | HLA_complex_group_2_pseudogene_7 | |||||
| 3.8-1.3 | ||||||
| LOC100507680 | HLA_class_I_histocompatibility_antigen,_A-69_alpha_chain-like_isoform_13 | |||||
| HLA-W | ||||||
| HCG2P6 | ||||||
| MICD | ||||||
| HCG9 | HLA_complex_group_9 | |||||